Figures & data
Summary of the bat-CoVs detection in bats from the Zhejiang province of China
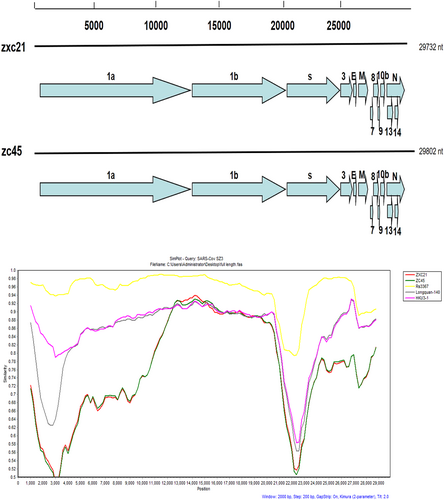
Similarity plots were conducted with SARS CoV SZ3 as the query and bat SL-CoVs, including Rs3367, Longquan-140, and HKU3-1, as potential parental sequences. The analysis was performed using the Kimura model, with a window size of 2000 base pairs and a step size of 200 base pairs
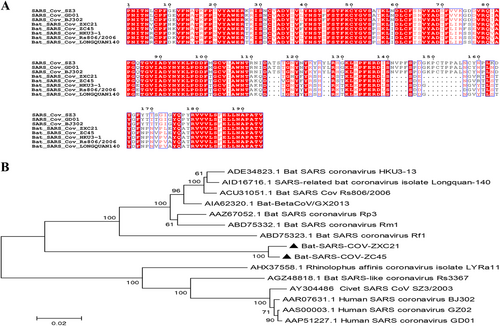
a Amino acid sequence comparison of the S1 subunit. The receptor-binding domain (aa 318–510) of SARS-CoV. b A phylogenetic analysis of the entire S1 amino acid sequences based on the neighbor-joining method. The SARS-CoV-GD01, BJ302, and GZ02 strains were isolated from patients of the SARS outbreak in 2003. The SARS-CoV SZ3 was identified from civets in 2003. Other bat-SL-CoVs were identified from bats in China.The sequences of SL-CoVs in this study are marked as filled triangles
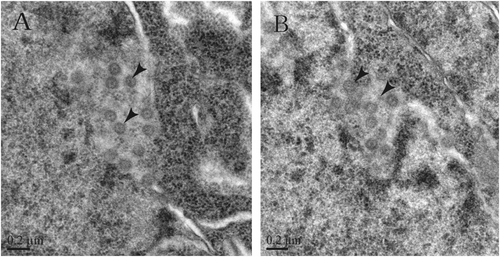
Sectioned brain, intestine, lung and liver tissues were sampled from rats infected with bat-SL-CoV ZC45
a, b CoV-like particles are considered SL-CoVs ZC45 in different locations of the infected rat brain tissues
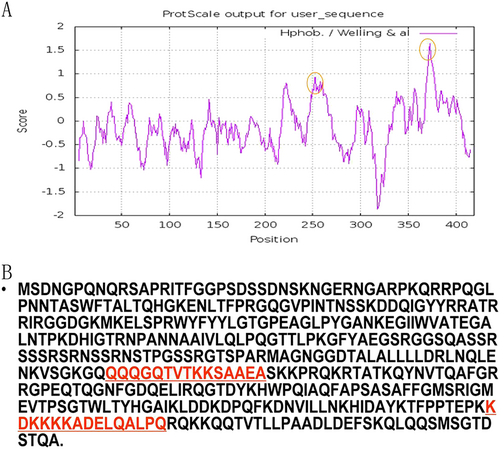
a The predicted antigenicity for the N protein. b Amino acid sequence of the N protein. The high antigenicity portion is indicated in the red circle. The two synthesized polypeptides are indicated in red
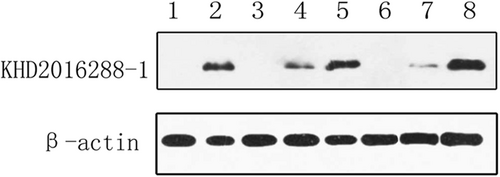
Proteins from the following tissues were analyzed: rat brain from the control specimen (lane 1), intestinal tissue from bat ZC45 (lane 2), intestinal tissue from the infected rat (lane 3,6), lung tissue from the infected rat (lane 4,7), and brain tissue from the infected rat (lane 5,8)