Figures & data
Fig. 1 Maximum likelihood phylogenetic tree based on the entire VP1 coding region sequences of EV-B80 available from GenBank. The four EV-B80 strains in this study are indicated by solid diamonds, and the prototype of EV-B80 is indicated by a solid circle. The branches are color-coded according to the location of sample collection (India = “green”, China = “red”). The scale bars indicate the substitutions per site per year. The numbers at the nodes indicate the bootstrap support for the node (percentage of 1000 bootstrap replicates)
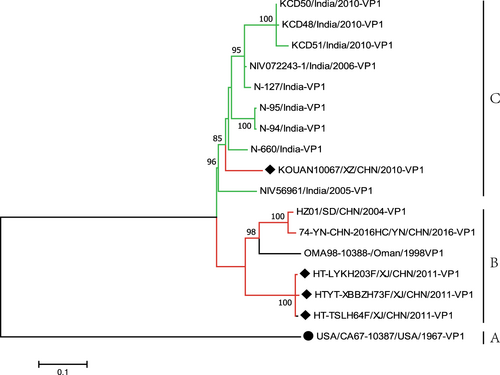
Fig. 2 The genomic map (upper) and recombination events predicted for the four EV-B80 strains. The likely backbone and other genetic components were identified based on Table S2. The genome of EV-B80 is shown as a black block. Genetic components identified by RDP4 that were involved in recombination events are shown as light gray blocks. Likely breakpoint positions are shown above the genome. a Recombination events of HT-LYKH203F; b recombination events of HT-TSLH64F; c recombination events of HTYT-XBBZH73F;d recombination events of KOUAN10067
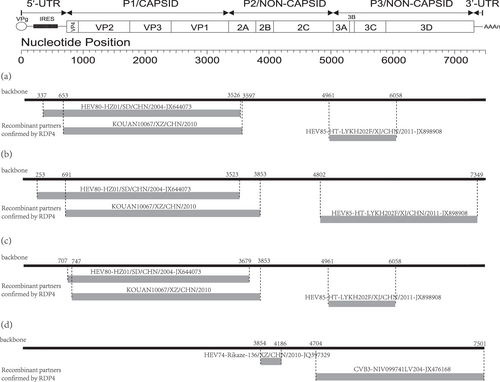
Fig. 3 a Similarity and bootscanning analysis of strain KOUAN10067 with CV-B3 strains isolated from India, the prototype of EV-B80, a field EV-B74 strain (Rikaze-136/XZ/CHN/2010), and a field EV-B80 strain (HZ01/SD/CHN/2004). Strain KOUAN10067 was used as a query sequence. b Midpoint-rooted maximum likelihood phylogenetic tree of isolates based on regions excluding P2 and P3 coding regions. c Midpoint-rooted maximum likelihood phylogenetic tree of isolates based on the P2 coding region. d Midpoint-rooted maximum likelihood phylogenetic tree of isolates based on the P3 coding region. All branches of the trees are colored according to the result of the bootscanning analysis except for strain KOUAN10067 (dark yellow). The sequences analyzed in the recombination and phylogenetic trees were identical
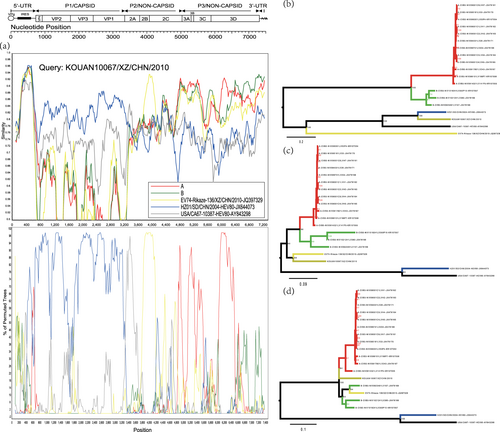
Fig. 4 a Similarity and bootscanning analysis of EV-B85, EV-B106, and EV-B80 strains with potential parents. The J group was used as a query sequence. b Midpoint-rooted maximum likelihood phylogenetic tree of isolates was constructed based on regions excluding the P2 and P3 coding regions. c Midpoint-rooted maximum likelihood phylogenetic tree of isolates based on the P2 coding region. d Midpoint-rooted maximum likelihood phylogenetic tree of isolates based on the P3 coding region. All branches of the trees are colored according to the result of bootscanning analysis except for J group (dark yellow). The sequences analyzed in recombination and phylogenetic trees were identical
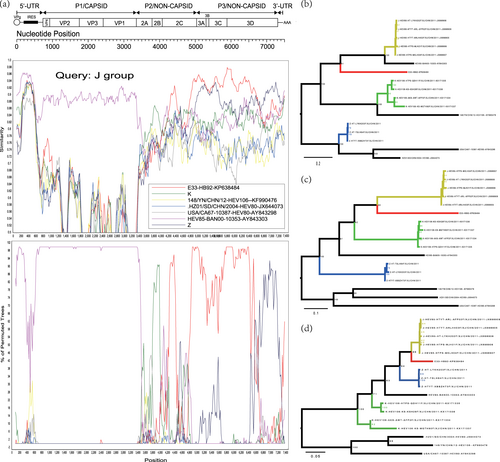
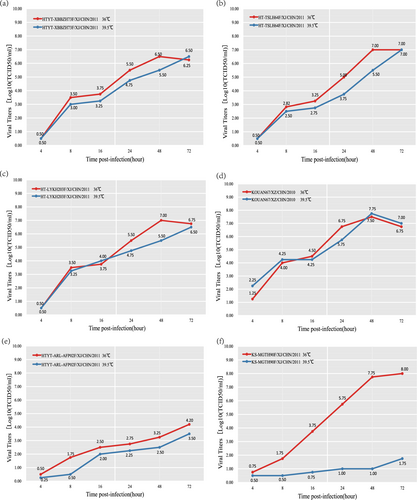
Fig. 5 Temperature sensitivity test curves of the four EV-B80 strains. Blue and red lines represent the growth trends of the viruses on RD cells at 36 and 39.5 °C, respectively. The Xinjiang EV-B85 strain (HTYT-ARL-AFP02F/XJ/CHN/2011, showing nontemperature sensitivity) and the EV-B106 strain (KS-MGTH90F/XJ/CHN/2011, showing temperature sensitivity) were used as experimental controls. a strain HTYT-XBBZH73F; b strain HT-TSLH64F; c strain HT-LYKH203F; d strain KOUAN10067e strain HTYT-ARL-AFP02F (EV-B85); f strain KS-MGTH90F (EV-B106)