Figures & data
Fig. 1 Illustration of a point null hypothesis, H0; the estimated effect that is the best supported hypothesis, ; the confidence interval (CI) for the estimated effect [
]; and the interval null hypothesis
.
![Fig. 1 Illustration of a point null hypothesis, H0; the estimated effect that is the best supported hypothesis, Ĥ=θ̂; the confidence interval (CI) for the estimated effect [CI−,CI+]; and the interval null hypothesis [H0−,H0+].](/cms/asset/85639327-eee8-42fb-8091-ab921fcdb23a/utas_a_1537893_f0001_c.jpg)
Table 1 Comparison between the classical and second-generation p-value.
Fig. 2 Manhattan plot of SNP associations, colored according to second-generation p-value status. The x-axis shows SNP position on chromosome 6 and the position of the organic cation transporter gene (SLC22A3). The top 16 associations by p-value rank are in green and the top 16 associations by second-generation p-value rank are in black. Bonferroni, FDR, and unadjusted significance levels are show by dashed lines.
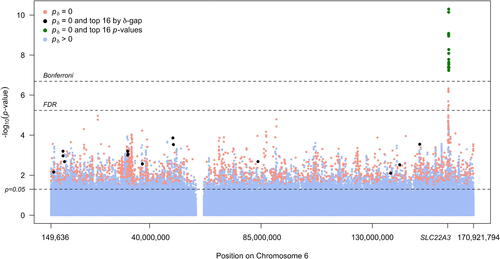
Fig. 3 Modified Manhattan-style plot of SNP associations as measured by the delta-gap. Points are colored according to second-generation p-value status. The x-axis shows SNP position on chromosome 6 and the position of the organic cation transporter gene (SLC22A3). The top 16 associations by p-value rank are in green and the top 16 associations by second-generation p-value rank are in black.
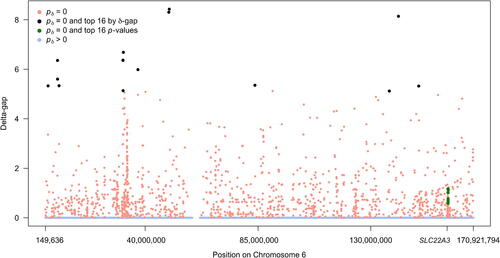
Fig. 4 1/8 likelihood SIs for the estimated odds ratio for SNPs with the smallest 500 p-values. The x-axis is p-value rank. The interval null hypothesis (in gray) ranges from 0.9 to 1.1111 (= 1/0.9). Intervals are colored according to second-generation p-value status. The top 16 Bonferroni SNPs are in green and the top 16 SGPV SNPs are in black. Bonferroni (“Bonf”) and FDR significance levels are shown by dashed lines.
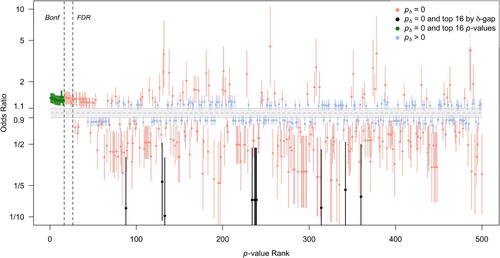
Fig. 5 Mirrored waterfall plot. 1/8 likelihood SIs for the estimated odds ratio for SNPs in the top 2,000 second-generation p-value ranking. The x-axis is SGPV rank. The interval null hypothesis (in gray) ranges from 0.9 to 1.1111 (= 1/0.9). Intervals are colored according to second-generation p-value status. The top 16 Bonferroni SNPs are in green and the top 16 SGPV SNPs are in black.
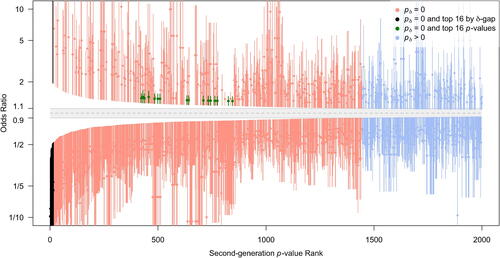
Fig. 6 Cross-tabulation of classical p-values and second-generation p-values in the prostate cancer data example.
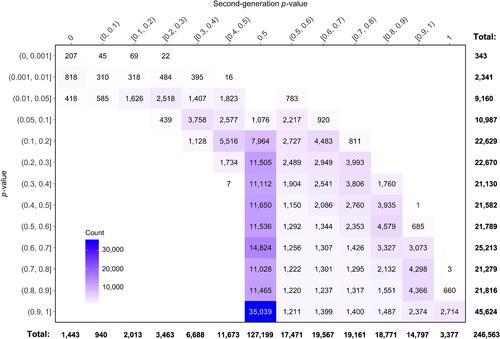
Table 2 False discovery rates of 5 SGPV = 0 findings computed under various null and alternative hypothesis configurations.
