Figures & data
Figure 1. Overview of nitrogen response in plants. Nitrogen source limitation (right half) leads to axial root elongation and root cortical aerenchyma formation. Nitrogen source limitation also increases ammonium and nitrate uptake activity. Supplementation of nitrogen nutrients (left half), either nitrate or ammonium, induces lateral root proliferation and increase of nitrate or ammonium uptake capacity. Note that the promotion of root cortical aerenchyma formation under nitrogen limitation has been reported only in rice and maize. The specific responses might differ among different plant species.
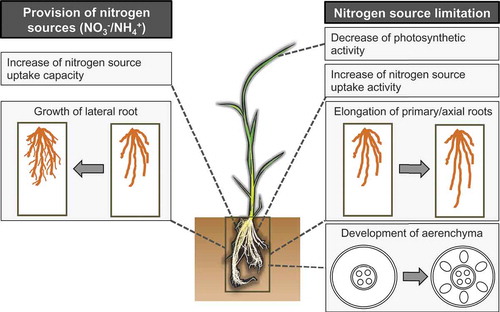
Figure 2. Importance of nitrate as a nitrogen source in lowland rice. (a) Hypothetical scheme of nitrate utilization in rice grown under flooded conditions. Oxygen is supplied to the root from the shoot through aerenchyma. Conveyed oxygen is diffused into the rhizosphere and produces locally aerobic conditions. Then, it activates bacteria involved in nitrification, and the microbes produce nitrate, which is immediately taken up by plants via nitrate transporters. (b) Selection for nitrate transporters (NRTs) during indica rice domestication and modern breeding. Plants displayed with NRT alleles with better function (hexagons) were selected over the unfavorable ones (triangles), possibly due to their effects on nitrogen utilization, biomass production, and grain yield.
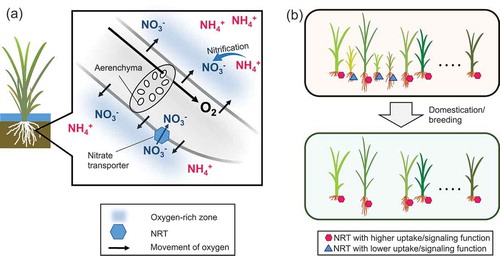
Figure 3. Molecular mechanism of nitrate-induced gene expression. (a) Structure of Arabidopsis NLP7. NLP proteins contain an amino-terminal conserved domain (indicated by dark gray), a DNA-binding RWP-RK domain, and a PB1 domain. The amino-terminal region to the RWP-RK domain is nitrate responsive. The conserved serine residue (Ser 205) of NLP7 is phosphorylated in response to nitrate supply. (b) Without nitrate, NLP7 is inactive and mostly resides in the cytosol due to active export out of the nucleus. Nitrate enters into cells through nitrate transporters (NRT1.1 and NRT2). Calcium-dependent protein kinases (CPK10, CPK30, and CPK32) are located at the plasma membrane, but nitrate supply stimulates their translocation to the nucleus and their activation through elevated cytosolic Ca2+. Ca2+-bound CPKs interact with NLP7 inside the nucleus and phosphorylate the conserved serine residue, leading to NLP7 accumulation in the nucleus. Activated NLP7 binds to the cognate DNA sequences (nitrate-responsive elements [NREs]) and promotes transcription of target genes, including NIA1 (nitrate reductase gene), NITR2 (plastidic nitrite transporter gene, Maeda et al. Citation2014), and NIR1 (nitrite reductase gene). The presence of a bifurcating pathway for nitrate signaling is also implicated.
![Figure 3. Molecular mechanism of nitrate-induced gene expression. (a) Structure of Arabidopsis NLP7. NLP proteins contain an amino-terminal conserved domain (indicated by dark gray), a DNA-binding RWP-RK domain, and a PB1 domain. The amino-terminal region to the RWP-RK domain is nitrate responsive. The conserved serine residue (Ser 205) of NLP7 is phosphorylated in response to nitrate supply. (b) Without nitrate, NLP7 is inactive and mostly resides in the cytosol due to active export out of the nucleus. Nitrate enters into cells through nitrate transporters (NRT1.1 and NRT2). Calcium-dependent protein kinases (CPK10, CPK30, and CPK32) are located at the plasma membrane, but nitrate supply stimulates their translocation to the nucleus and their activation through elevated cytosolic Ca2+. Ca2+-bound CPKs interact with NLP7 inside the nucleus and phosphorylate the conserved serine residue, leading to NLP7 accumulation in the nucleus. Activated NLP7 binds to the cognate DNA sequences (nitrate-responsive elements [NREs]) and promotes transcription of target genes, including NIA1 (nitrate reductase gene), NITR2 (plastidic nitrite transporter gene, Maeda et al. Citation2014), and NIR1 (nitrite reductase gene). The presence of a bifurcating pathway for nitrate signaling is also implicated.](/cms/asset/1fc7a11d-e1db-40a7-9eb2-3e184a1f47ba/tssp_a_1360128_f0003_oc.jpg)
