Figures & data
Figure 1. Concentration versus time profile of Compound 1 following 5mg/kg iv (red) and po (blue) in 10% DMSO, 1% tween 80 in saline. Profiles are from individual mice (n = 3). Analysis was carried out by LC-MS/MS with selected reaction monitoring. Clearance and bioavailability have been calculated with Phoenix non-compartmental analysis.
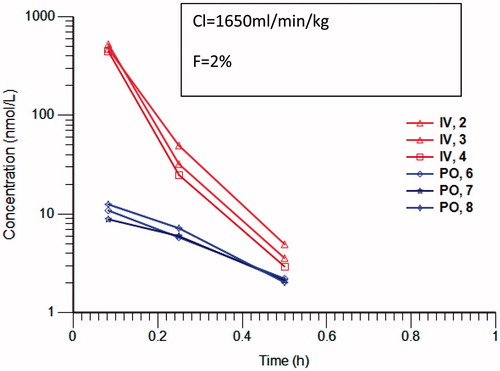
Figure 2. Fragmentation pattern of Compound 1 (m/z 379.17) and its oxidation product (m/z 395.17). The core pyrido[3,4-d]pyrimidin-4(3H)-one fragment in 1 (m/z 146.04) is oxidised in the metabolite (m/z 162.03). No further fragmentation was observed.
![Figure 2. Fragmentation pattern of Compound 1 (m/z 379.17) and its oxidation product (m/z 395.17). The core pyrido[3,4-d]pyrimidin-4(3H)-one fragment in 1 (m/z 146.04) is oxidised in the metabolite (m/z 162.03). No further fragmentation was observed.](/cms/asset/7c387135-3dd0-4765-85c9-3a96a252fc22/ixen_a_1230245_f0002_c.jpg)
Figure 3. 1H NMR spectrum of compound 1 and its oxidation product referenced to internal deuterated solvent. Protons C5 and C6 are still present in the metabolite suggesting that the oxidation is at position C2 of the pyrido[3,4-d]pyrimidin-4(3H)-one scaffold.
![Figure 3. 1H NMR spectrum of compound 1 and its oxidation product referenced to internal deuterated solvent. Protons C5 and C6 are still present in the metabolite suggesting that the oxidation is at position C2 of the pyrido[3,4-d]pyrimidin-4(3H)-one scaffold.](/cms/asset/849692be-c4ad-464f-8aac-704e7903f72e/ixen_a_1230245_f0003_b.jpg)
Figure 4. Structure of a number of pyrido[3,4-d]pyrimidin-4(3H)-one derivatives and clearance in mouse and human cytosol following incubation of 1μM compound in the absence and presence of the aldehyde oxidase inhibitor raloxifene. Values are means ± SD of n = 3 replicate analysis. Statistical analysis carried out with paired t test comparing clearance with and without inhibitor. ***p < 0.001.
![Figure 4. Structure of a number of pyrido[3,4-d]pyrimidin-4(3H)-one derivatives and clearance in mouse and human cytosol following incubation of 1μM compound in the absence and presence of the aldehyde oxidase inhibitor raloxifene. Values are means ± SD of n = 3 replicate analysis. Statistical analysis carried out with paired t test comparing clearance with and without inhibitor. ***p < 0.001.](/cms/asset/8e725f17-541a-4428-a6b6-8705b0a8a10b/ixen_a_1230245_f0004_b.jpg)
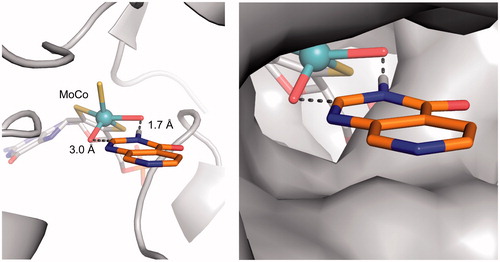
Figure 5. Proposed binding mode of compound 3 (orange sticks) in the human hAOX1 substrate binding site (PDB code 4uhw, grey). Cartoon representation of the protein with the proposed binding mode of 3 to the metal-coordinating oxygen atoms within the molybdenum cofactor MoCo (left); Surface representation illustrating the proposed binding mode (right).