Figures & data
Table 1. Conditions for CYP-Silensomes incubations.
Table 2. Characteristics of the mechanism-based inhibitors selected.
Table 3. Estimated CYP contribution.
Figure 1. Specificity of the inhibition of CYP activities in CYP1A2-, CYP2B6-, CYP2C8-, CYP2C9-, CYP2D6-Silensomes at initial rate (Km) and saturating conditions (Vmax) of substrates. CYP activities at Km and Vmax concentration of substrates in CYP1A2-Silensomes (A, B), CYP2B6-Silensomes (C, D), CYP2C8-Silensomes (E, F), CYP2C9-Silensomes (G, H), CYP2D6-Silensomes (I, J), and corresponding control-Silensomes were compared using the following CYP probe substrates: phenacetin for CYP1A2, bupropion for CYP2B6, paclitaxel or amodiaquine for CYP2C8, diclofenac for CYP2C9, omeprazole or S-mephenytoin for CYP2C19, dextromethorphan for CYP2D6, chlorzoxazone for CYP2E1 and midazolam (a), nifedipine (b) and testosterone (c) for CYP3A4 used at either concentrations close to Km (non-saturating conditions) and Vmax (saturating conditions) (see details in ). Inhibition is expressed as a % activity in control-Silensomes, mean ± SEM of experiments performed in triplicate on two CYP-Silensomes batches (saturating and non-saturating conditions) except CYP1A2 (three batches at non-saturating conditions) and CYP2B6 (one batch at non-saturating conditions), independent CYP-Silensomes batches in triplicate. (1) is the result of Anova tests comparing non-target CYP inhibition to be different from target CYP inhibition; (2) corresponds to statistically significant inhibition different from 0 (ns 0 = non significantly different from 0).
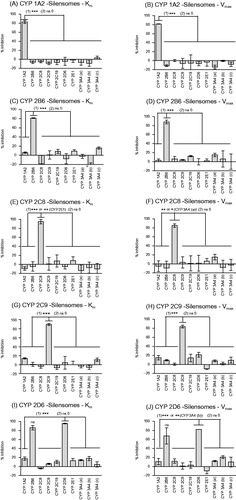
Figure 2. Correlation between observed in vivo and predicted contribution to drug metabolism using a direct quantitative method with CYP-Silensomes (A) or indirect rhCYP approach with the relative activity factors (RAFs) using nifedipine (B) and testosterone (C) as substrates for the CYP3A4 activity. All contribution values are shown in . The solid line indicates the line of perfect correlation and the dotted line ± 15% error interval.
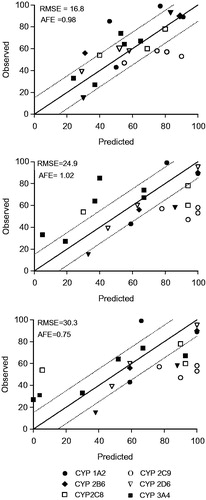
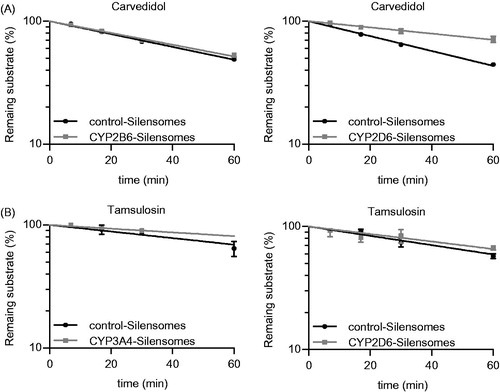
Figure 3. Determination of the fmCYP2D6 of Carvedidol (A) and both fmCYP2D6 and fmCYP3A4 of tamsulosin (B) using CYP2D6-, CYP2B6- (A, B) and CYP3A4-Silensomes (B) and their respective homologous control-Silensomes. Drugs were incubated at 0.1 µM and the disappearance of the parent compound was measured over 60 min. Percentages of remaining substrates are mean ± SEM of three independent experiments and were linearly fitted to determine clearance.