Figures & data
Figure 1. Location of the 13 sampling sites along the continuum of the Pine River. Sampling site numbers correspond to and . White line approximates the border of the Pine River watershed. Base map © Google, TerraMetrics.
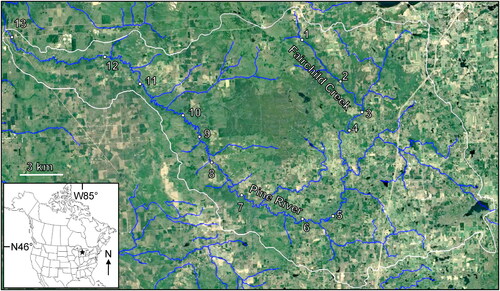
Table 1. Each site’s position from upstream (1) to downstream (13), GPS coordinates, and the number of samples collected at that site. Site numbers are in .
Table 2. Mean specimen abundance values for each of the 50 EPT taxa caught in this study, rounded to the nearest whole number. Site numbers correspond to and .
Figure 2. NMDS ordination of the 13 sampling sites based on mean EPT FFG biomass per site. P-values from a Monte Carlo test of non-random ordination structure. Arrow lengths denote strength of correspondence of FFG biomass with axis 1 values, only r2 > 0.50 shown. N = 3–4 per site ().
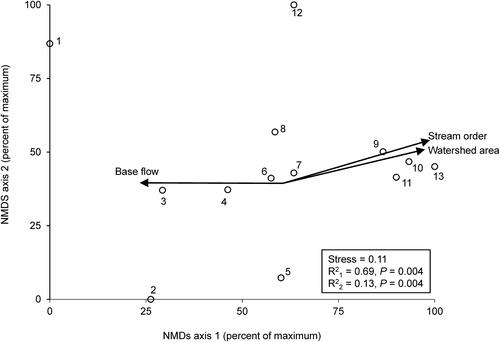
Figure 3. Second degree polynomial regression models of mean (±SE) AFDM (dependent variable) based on axis 1 scores from (independent variable) for filtering collectors (A), gathering collectors (B), predators (C), scrapers (D), shredders (E). Summary equations and R2 values in separate box. Each marker represents the mean (±SE) of 1 of the 13 sampled sites. N = 3–4 per site.
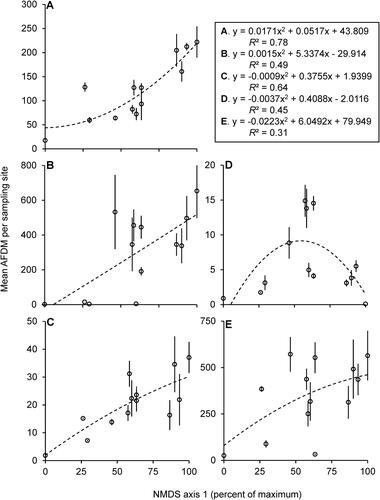
Figure 4. Second degree polynomial regression model of mean (±SE) number of EPT genera caught (dependent variable) based on axis 1 scores from (independent variable) for the 13 sampling sites, with summary equation and R2 values. Each marker represents the mean (±SE) of 1 of the 13 sampled sites. N = 3–4 per site.
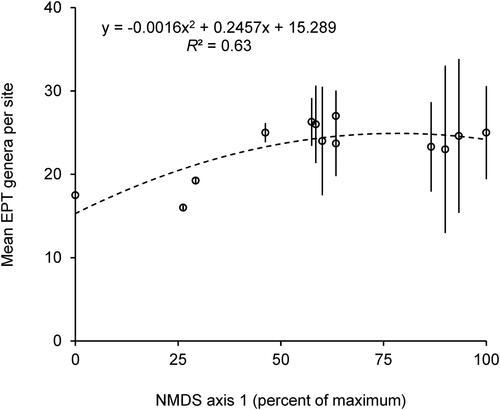
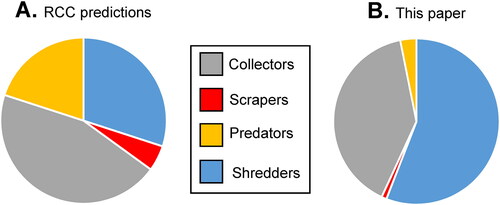
Figure 5. Approximate relative biomass of each FFG as predicted by Vannote et al. (Citation1980) for 1st–4th order streams (A), compared to the data in our study when expressed as relative determined biomass (B).
Data availability statement
Data are available on the Open Science Framework site: https://osf.io/nqfdh/.
