Figures & data
Figure 1. Site locations and discussed in the text. Teotihuacan city, Templo de Quetzalcoatl, and La Ventilla (this study). Paquimé, Jicaro, Convento (Morales-Arce et al. Citation2017), Tlateloco (Morales-Arce et al. Citation2019). Zapotec, Mazahua (Mizuno et al. Citation2014), Maya (Mizuno et al. Citation2017) were present-day Native Mesoamerican individuals. Teotihuacan city, Templo de Quetzalcoatl, La Ventilla, Paquimé, Jicaro, Convento, and Tlateloco were ancient individuals.
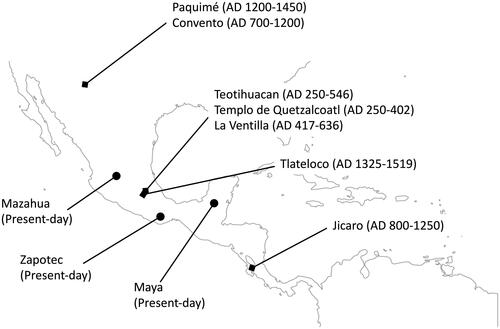
Table 1. NGS results of mitogenome of Teotihuacan city, Templo de Quetzalcoatl, and LaVentilla individuals.
Figure 2. Probability distributions for the calibrated dates of six collagen samples from Teotihuacan city, Templo de Quetzalcoatl, and La Ventilla. The plus mark indicates the median of the each.
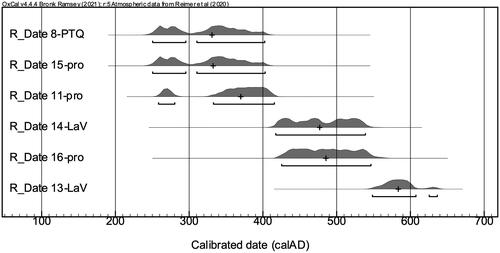
Figure 3. Relationship between δ13C and δ15N for six ancient samples from Teotihuacan city, Templo de Quetzalcoatl, and La Ventilla sites (this study) together with ancient Mesoamerican stable isotope analysis results. Black circle for Teotihuacan, black rhombus for Templo de Quetzalcoatl, and black triangle for La Ventilla. Grey circle, Square, Triangle were pre-maize diet, Transitional maize diet, and staple maize diet, respectively. These isotopic values from Classic Period maize agriculturalists from across the Maya lowlands (Kennett et al. Citation2020).
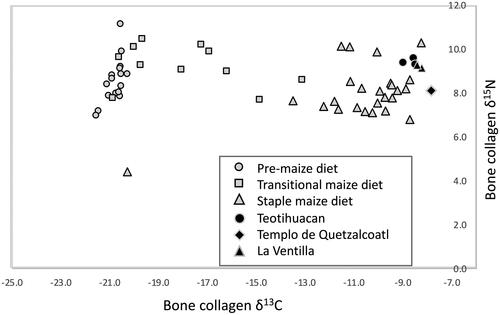
Figure 4. The median-joining network of mitogenome sequences for 13 ancient and 122 present-day Mesoamericans. The size of the circle reflects the number of individuals. Dark purple: ancient sequences belong to Teotihuacan city, Templo de Quetzalcoatl, and La Ventilla, Paquimé, Jicaro, Convento, and Tlateloco. Green: present-day Maya. Mazanta: present-day Mazahua. Blue: present-day Zapotec. Grey circles delinate mitogenome haplogroups: A2, B2, C1, D1, and D4.
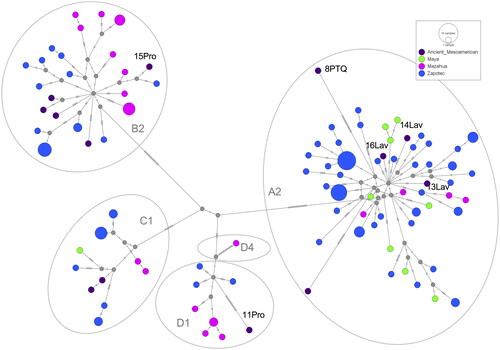
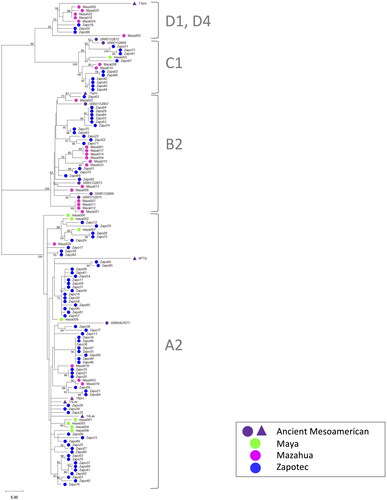
Figure 5. Maximum Parsimony tree of whole mitogenomes for 13 ancient and 122 present-day Mesoamericans. One of the most parsimonious trees (length = 491) is shown. The consistency index is 0.835031 (0.714789), the retention index is 0.963431 (0.963431), and the composite index is 0.804494 (0.688650) for all sites and parsimony-informative sites (in parentheses). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates and higher than 50%) are shown next to the branches. Dark purple: ancient Mesoamericans including Teotihuacan city, Templo de Quetzalcoatl, and La Ventilla, Paquimé, Jicaro, Convento, and Tlateloco. Green: present-day Maya. Magenta: present-day Mazahua. Blue: present-day Zapotec. Mitogenome haplogroups A2, B2, C1, D1, and D4 were assigned. The other possible trees equally parsimonious are shown in Figure S2.
Supplemental Material
Download Zip (3.8 MB)Data availability statement
Sequencing data produced in this study have been deposited in the GenBank (under accession numbers LC726583–LC726588) for ancient samples.
