Figures & data
Figure 1. Overview of the Mendelian randomisation design.
The solid lines represent meaningful pathways, and the dashed lines represent nonexistent pathways in Mendelian randomisation analysis.
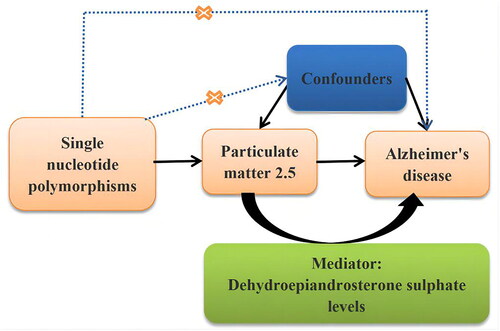
Table 1. Summary of the included traits in the Mendelian randomisation study.
Table 2. Detailed information on genetic instruments associated with Alzheimer’s disease.
Figure 2. Association of the genetically determined PM2.5 and dehydroepiandrosterone sulphate with Alzheimer’s disease.
CI: confidence interval; OR: odds ratio; PM: particulate matter; DHEAS: dehydroepiandrosterone sulphate; MR-PRESSO: Mendelian randomisation pleiotropy residual sum and outlie; MVMR: multivariable Mendelian randomisation.
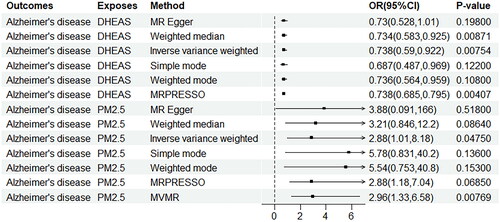
Figure 3. Forest plot of PM2.5 and dehydroepiandrosterone sulphate associated with risk of Alzheimer’s disease. The x-axis showed the MR effect size for PM2.5 and dehydroepiandrosterone sulphate on Alzheimer’s disease: (A)PM 2.5, (B) dehydroepiandrosterone sulphate. The y-axis showed the analysis for each of the SNPs.
PM: particulate matter; SNP: single nucleotide polymorphism; MR: Mendelian randomisation.
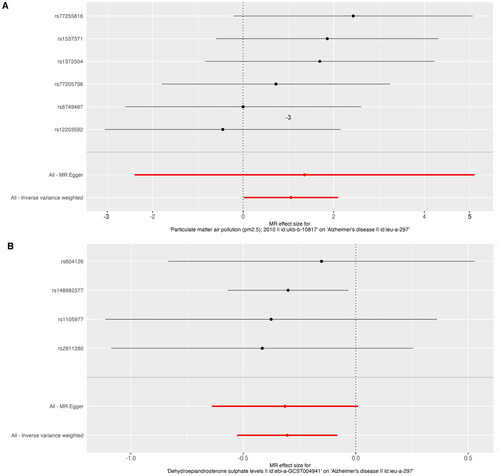
Table 3. Mendelian randomisation assessing the causal association between expose/mediator and Alzheimer’s disease.
Table 4. Mediation analysis of the mediation effect of PM2.5 on Alzheimer’s disease via dehydroepiandrosterone sulphate levels.
Figure 4. A scatter plot is used to visualise the causal effect association of PM2.5 and dehydroepiandrosterone sulphate level with Alzheimer’s disease: (A)PM 2.5, (B) dehydroepiandrosterone sulphate. The x-axis showed the SNP effect and SE on each PM2.5 anddehydroepiandrosterone sulphate. The y-axis showed the SNP effect and SE on Alzheimer’s disease.
PM: particulate matter; SNP: single nucleotide polymorphism; SE: standard error.
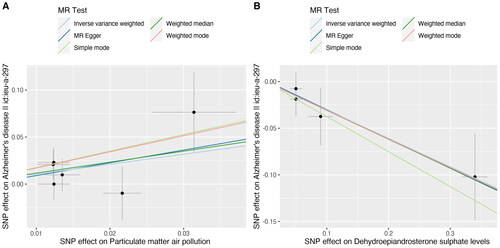
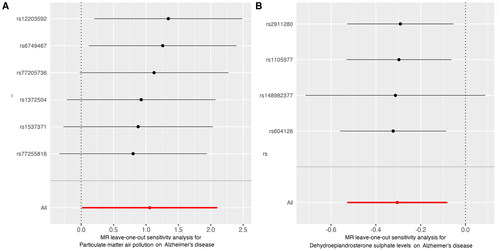
Figure 5. Leave-one-out sensitivity analysis for the effect of PM2.5 and dehydroepiandrosterone sulphate on Alzheimer’s disease: (A)PM 2.5, (B) dehydroepiandrosterone sulphate. The x-axis showed MR leave-one-out sensitivity analysis for PM2.5 or dehydroepiandrosterone sulphate on Alzheimer’s disease. The y-axis showed the analysis for the effect of leave-one-out of SNPs on Alzheimer’s disease.
MR: Mendelian randomisation; PM: particulate matter; SNP: single nucleotide polymorphism.
Availability of data and materials
The GWAS dataset were publicly available from the IEU open GWAS database (https://gwas.mrcieu.ac.uk/). The functions used in the TwoSampleMR package are available from the website https://mrcieu.github.io/TwoSampleMR/.
