Figures & data
Table 1. Verticillium dahliae isolates collected from olive tree (Olea europea) in Tunisia.
Table 2. List of amplified genomic regions and primers.
Fig. 1 (Colour online) Morphological characteristics of different V. dahliae morphotypes cultured on PDA for 15 days at 25 °C. A: Morphotype 1 with milky white and flocculose mycelium; B: Morphotype 2 with greyish-white surface; C: Morphology of conidiophores, phialides and conidia of V. dahliae; D: pure culture of V. dahliae.
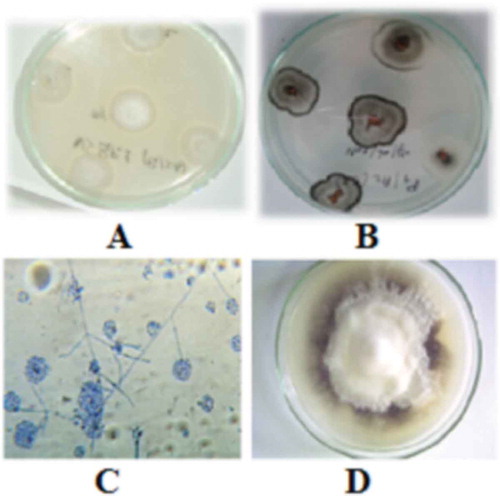
Fig. 2 Gel electrophoresis of the amplified rDNA internal transcribed spacer (ITS) specific region of Verticillium dahliae isolates. Lanes 1–16 are loaded as follows: DNA size marker, PCR products of Vsb 1.2, VSBP7, Vms 3.3, Vmh3.2, VK1.11, Vk1.3, Vcm2, V.hmd, VSB1.1, V30, Vd4, Vz17, VS 1.1G, Vms 3.2, and negative control.
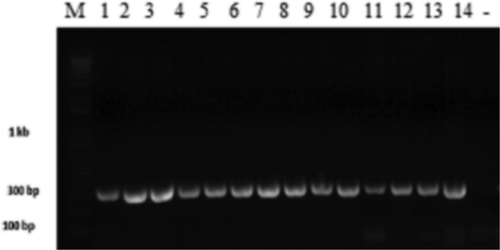
Fig. 3 Disease severity (DS) of V. dahliae isolates recovered from olive trees in Tunisia (histograms denoted by the same letter are not significantly different according to LSD test; α ≤ 0.05).
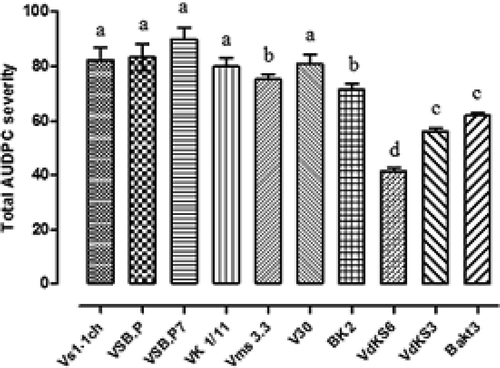
Table 3. Banding patterns of the amplified rDNA intergenic spacer (IGS) region after digestion with RsaI and EcoRI.
Fig. 4 Random amplified microsatellite (RAMS) pattern of Verticillium dahliae isolates using CGA primer. Lanes 1–15 are loaded with PCR products of VDKS5, Vd4, BAKT3, VS3.3F, VS1.2 T, VMS3.2, VMS2, VDKS6, VDKS3, VK1.2, Vd2, VK6.11, VFA2, BAKT11 and BAKT7, respectively. DNA size marker is indicated by M.
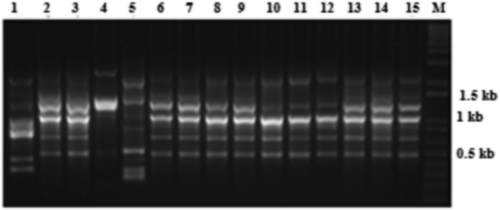
Fig. 5 Random amplified microsatellite (RAMS) pattern of Verticillium isolates using CCA primer. Lanes 1–21 are loaded with PCR products of Vsh1.2, VSBP7, BK2, Vmh3.2, VK1.11, Vk1.3, Vcm2, VS1.1ch, VSB1.1, Vsh, V30, Vd3, Vd4, Vz17, VSB1.4, VHmd, Vd2, VDOR, VS1.2ch, VS1.1 G and BAKT7, respectively. DNA size marker is indicated by M.
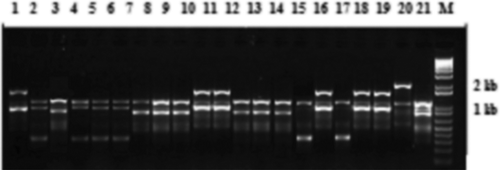
Table 4. RAMS banding patterns of the tested V. dahliae isolates using primers GT, CCA and CGA.
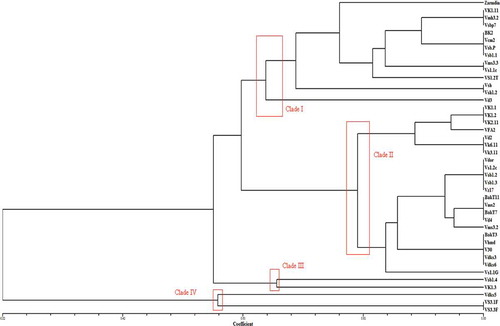
Fig. 6 (Colour online) Phylogenetic tree using matching coefficient based on RAMS bands of Verticillium fungal isolates with primer GT, CCA and CGA.