Figures & data
Fig. 1 Map of the counties surveyed for bacterial spot in processing tomato, 2010–2020. Northwest Ohio: Ottawa, Erie, Sandusky, Fulton, Wood, Henry, Putnam and Hancock; Southeast Michigan: Lenawee; and Central Indiana: Madison, Tipton counties.
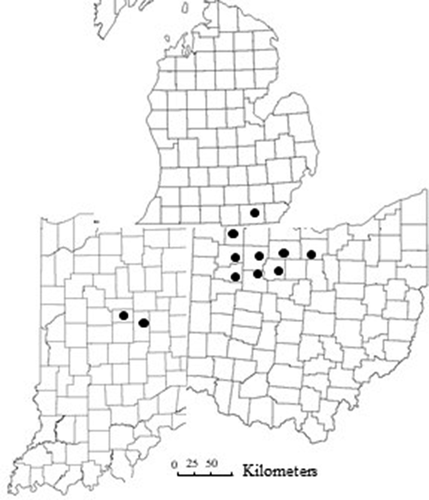
Table 1. Xanthomonas strains isolated from processing tomatoes in the U. S. Midwest, by species and geographical location, 2010–2020. Xanthomonas spp. identification is based on the qPCR outcome.
Fig. 2 BOX-PCR fingerprints of Xanthomonas spp. strains isolated from processing tomato in the U.S. Midwest. L) ladder 2 kb; 1 to 12) X. hortorum pv. gardneri; 13 and 14) putative X. perforans; 15 and 16) X. perforans; reference strains 17) X. hortorum pv. gardneri XcgA2; 18) X. euvesicatoria 110C; 19) X. perforans 1220.
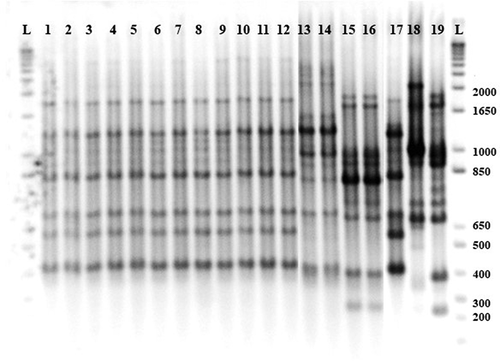
Fig. 3 Xanthomonas species distribution (percent) in processing tomatoes in Ohio, Indiana and Michigan, 2010–2020. Data for years 2012 and 2013 are not reported in the figure because of their small sample size, and no sampling was done in 2014–2016.
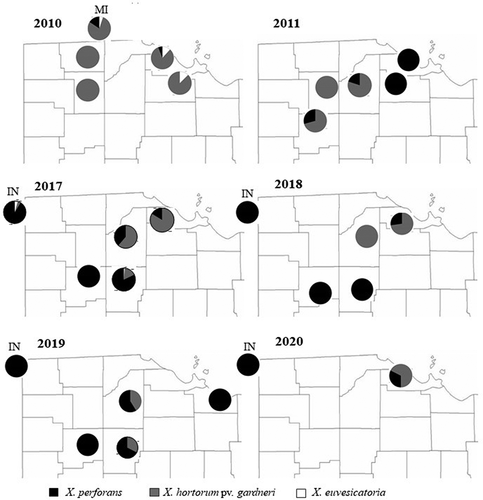
Table 2. Association between Xanthomonas species isolated from processing tomatoes in the U.S. Midwest and tomato seed sources.
Table 3. Association of Xanthomonas hortorum pv. gardneri and X. perforans with green and ripe processing tomato fruit, 2017–2020.
Table 4. Percentage of copper sulphate- and streptomycin sulphate-resistant Xanthomonas hortorum pv. gardneri and X. perforans strains isolated from processing tomatoes in the U.S. Midwest, 2010–2020.
Fig. 4 Mosaic charts showing the proportion of Xanthomonas spp. strains from processing tomatoes in the U.S. Midwest resistant to copper sulphate (200 µg mL−1) for each surveyed year. Resistance is indicated in black, sensitivity in white. The width of each column is proportional to the relative abundance of each species.
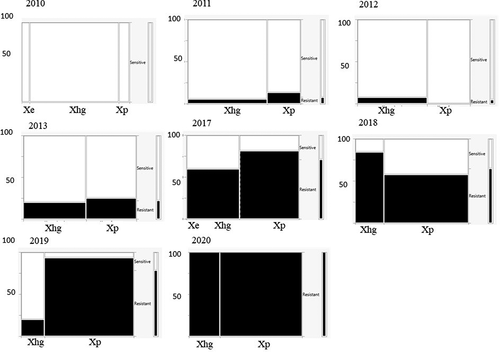
Fig. 5 Mosaic charts showing the proportion of Xanthomonas spp. strains from processing tomatoes in the U.S. Midwest resistant to streptomycin sulphate (200 µg mL−1) for each surveyed year. Resistance is indicated in black, sensitivity in white. The width of each column is proportional to the relative abundance of each species.
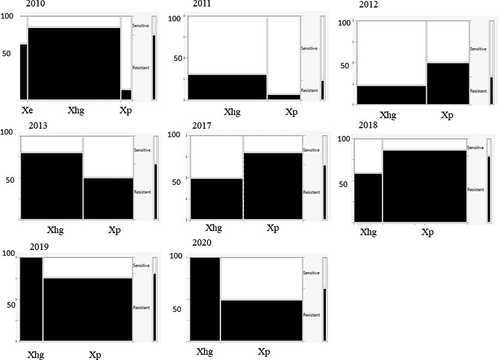
Table 5. Percentage of bacteriocin-producing Xanthomonas perforans strains isolated from processing tomatoes in the U.S. Midwest, 2010–2020 that inhibited the growth of X. euvesicatoria 91–106 K.
