Figures & data
Figure 1. Sir2/SIRT1 and signaling pathways in different species. In yeast Sir2p activity is increased through increased expression of the NAD+ salvage pathway enzyme, nicotinamidase PNC1, but also by increasing the NAD+/NADH ratio (or lowering NADH levels) in response to calorie restriction (CR). Mutation of the glucose sensing receptors Gpr1 and Gpa2 mimics the CR effects on Sir2p, which inhibit the formation of the deleterious extrachromosomal circular DNA repeats (ERCs) and contribute to life span extension in yeast. The glucose and nutrients fermentation pathway activates the AKT‐related kinase Sch9 which induces oxidative stress and participates in yeast ageing. Inactivation of this pathway promotes longevity in yeast. The longevity pathway is conserved amongst worms, flies and mammals. Activation of the insulin growth factor receptor IGFR (Daf‐2/dIGFR) activates the insulin receptor substrate IRS (p65/CHICO), which stimulates phosphoinositol‐3 kinase PI3K (AGE‐1/dPI3K), which in turn phosphorylates the PKB/AKT kinase. AKT phosphorylates and inactivates FOXO (Daf‐16/dFOXO). Inactivation of the insulin receptor pathway promotes longevity in worms, drosophila, and mammals through increased activity of FOXO. Sir2/SIRT1 activation induces life span extension through interaction with Daf‐16/FOXO factors. Gpr‐1 = G‐protein coupled receptor‐1; Gpa‐2 = G‐alpha protein‐2; FGM = Fermentable growth medium; PNC1 = pyrazinamidase/Nicotinamidase 1; Sir2p = yeast silent information regulator 2; NADH = reduced form of nicotinamide adenine dinucleotide; Sch9 = Saccharomyces cerevisiae protein kinase 9; ERCs = extrachromosomal circular DNA repeats; Daf‐16 = ‘dauer’ larvae transcription factor‐16; AGE‐1 = worm homolog of the phosphoinositol‐3 kinase; AKT = protein kinase B; Rpd3 = reduced potassium dependence 3 (class II histone deacetylase); IGF‐1 = insulin growth factor‐1; FOXO = forkhead box subgroup ‘O’ transcription factor; IRS = insulin receptor substrate; CHICO = drosophila homolog of IRS; SIRT1 = sirtuin 1.
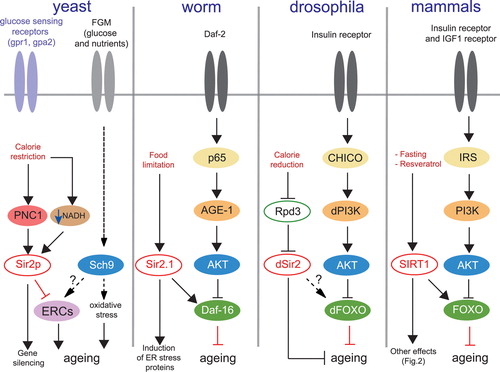
Figure 2. A: yeast and mammalian NAD+ salvage pathways. Yeast (red) Sir2p utilizes the cofactor NAD+ to deacetylate proteins and in this reaction produces nicotinamide (NAM) and O‐acetyl‐ADP‐ribose (O‐AA‐ribose). NAM is deaminated by the nicotinamidase PNC1 to form nicotinic acid (NA). NA will give rise successively to nicotinate mononucleotide (NAMN), nicotinate adenine dinucleotide (NAAD) and nicotinamide adenine dinucleotide (NAD+) by the enzymes nicotinate phosphoribosyl‐transferase (NPT1), nicotinate mononucleotide adenyltransferase (NMAT), and nicotinamide adenine dinucleotide (NAD) synthetase (QNS), respectively (red arrows). In mammals (blue) NAM is recycled to NAD+ in two steps through the formation of nicotinamide mononucleotide (NMN) by means of the NAM phosphoribosyltransferase NamPT (PBEF/visfatin) and nicotinamide mononucleotide adenyltransferase (NMNAT) (blue arrows). B: SIRT1 protective functions in metabolism and diseases. SIRT1 can be regulated positively by CR and SIRT‐activating compounds and negatively by SIRT inhibitors. SIRT1 activation induces survival of cardiomyocytes, protects neurons from cell death, and favors insulin secretion by repressing the uncoupling protein 2 (UCP2). SIRT1 decreases white adipocyte tissue formation through repression of PPARγ and promotes gluconeogenesis in response to fasting through PGC‐1α, and stimulates mitochondrial biogenesis in the brown adipose tissue (BAT) and the muscle through activation of PGC‐1α. NA = nicotinic acid; NAD+ = nicotinamide adenine dinucleotide; NAM = nicotinamide; NMN = nicotinamide mononucleotide; PNC1 = pyrazinamidase/Nicotinamidase 1; NamPT1 = nicotinamide phosphoribosyl transferase 1; PBEF = pre‐B cell enhancing factor; NPT1 = nicotinate phosphoribosyl transferase 1; NMAT1/2 = nicotinate mononucleotide adenyltransferase 1/2; QNS = NAD synthetase; NMNAT1 = nicotinamide mononucleotide adenyltransferase; O‐AA‐ribose = O‐acetyl‐ADP‐ribose; UCP‐2 = uncoupling protein 2; NFκB = nuclear factor kappa B; PPARγ = peroxisome proliferator‐activated receptor gamma; PGC‐1α = peroxisome proliferator‐activated receptor gamma coactivator‐1 alpha; BAT = brown adipose tissue; SIRT1 = sirtuin 1.
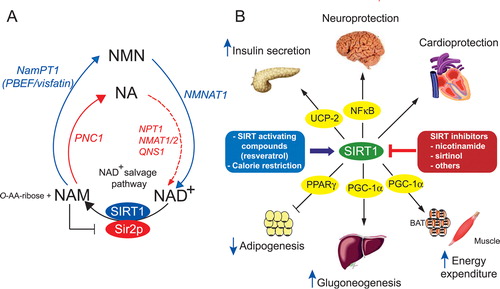
Table I. Expression pattern, cellular distribution, and function of sirtuin deacetylases. Sirtuins are differentially expressed in different organs based on their transcripts.