Figures & data
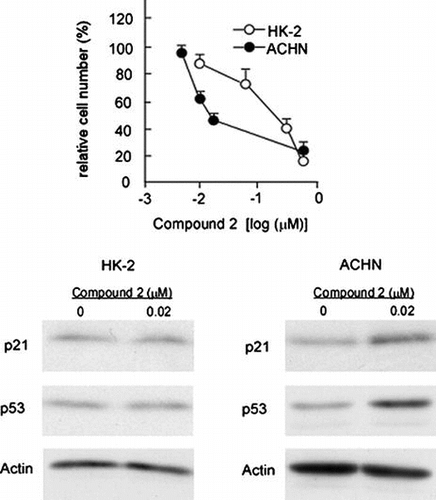
Fig. 1. Structure and cytotoxic activity of compounds 2, 9, 11, 12.
Notes: (A) Structures of 2, 9, 11, and 12. (B) IC50 values of 2, 9, 11, 12, etoposide, and cisplatin against HL-60 cells were posted from our paper (that of etoposide and cisplatin: as positive control) (1).
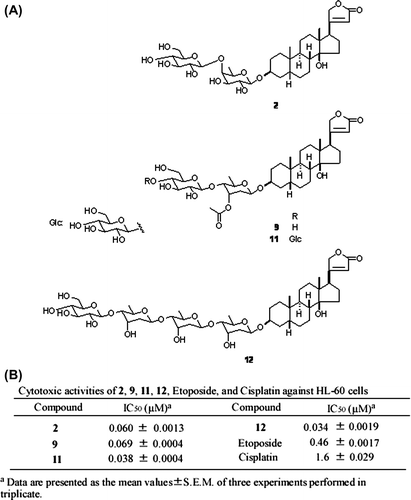
Fig. 2. Dose-dependent cytotoxicity of compounds 2, 9, 11, and 12 on HK-2, ACHN, Fa2 N-4, and HepG2 cells.
Notes: (A) HK-2 and ACHN cells were subjected to the treatment with cardenolide glycosides (2, 9, 11, and 12) and their cytotoxic effects were examined as described in Materials and methods. (B) Fa2 N-4 and HepG2 cells were subjected to the treatment with compound 2 and their cytotoxic effects were examined as described in Materials and methods. Data are presented as the mean values ± SEM of three experiments performed in triplicate.
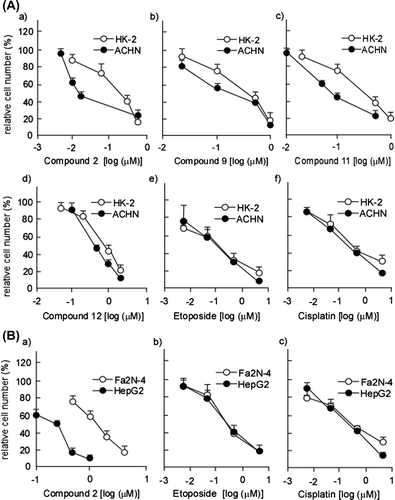
Table 1. Cytotoxic activities of 2, 9, 11, 12 against HK-2 and ACHN cells.
Fig. 3. Compound 2 does not cause morphological changes of nucleus.
Notes: Morphological changes in the nuclei of HK-2 and ACHN cells treated or left untreated with 2 were examined as described in Materials and methods (scale bar, 30 μm). Actinomycin D was used as positive control.
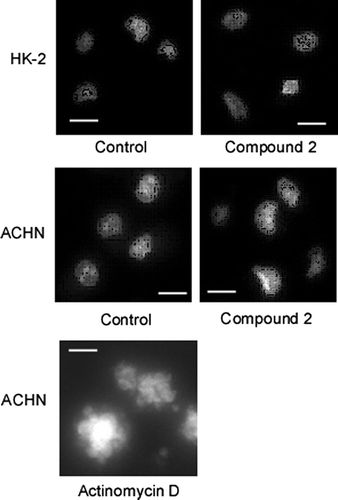
Table 2. Cytotoxic activities of 2 against Fa2 N-4 and HepG2 cells.
Fig. 4. p21 upregulation is responsible for compound 2-mediated cytotoxicity on ACHN Cells.
Notes: (A) HK-2 and ACHN cells were seeded at a density of 4.0 × 104 cells/35-mm dish and treated with control or p21/Cip1 siRNA (100 nM) using HiPerfect transfection reagent (Qiagen). After 72 h, total RNA was extracted and quantification of p21/Cip1 mRNA was performed as described in Materials and methods. Data are presented as the mean values ± SEM of three experiments performed in triplicate and analyzed via a Student’s t-test. (B) HK-2 and ACHN cells were treated with control or p21/Cip1 siRNA as described in (A). After 72 h, cell number/dish was determined. Data are presented as the mean values ± SEM of three experiments performed in triplicate and analyzed via a Student’s t-test. (C) ACHN cells were treated with control or p21/Cip1 siRNA as described in (A). After 24 h, cells were treated or left untreated with 2 (0.02 μM) for 72 h. Quantification of p21/Cip1 mRNA was performed as described in (A). (D) ACHN cells were treated with control or p21/Cip1 siRNA, and then subjected to the compound 2 treatment as described in (C). Then cell number/dish was determined as described in (B).
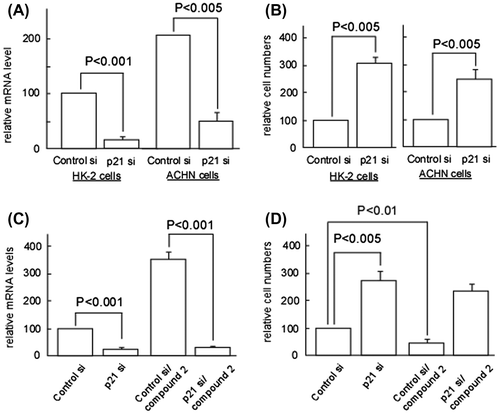
Fig. 5. Analyses for p21/Cip1, p16/INK4a, and FXR expressions in 2-treated HK-2, ACHN, Fa2 N-4, and HepG2 Cells.
Notes: HK-2 and ACHN seeded at a density of 8.0 × 104 cells/35-mm dish and Fa2 N-4 and HepG2 cells seeded at a density of 4.0 × 104 cells/35-mm were treated with various concentrations of 2. After 48 h (Fa2 N-4 and HepG2 cells) or 72 h (HK-2 and ACHN cells), total RNA was extracted and quantification of p21/Cip1, p16/INK4a, and FXR mRNAs was performed as described in Materials and methods. Data are presented as the mean values ± SEM of three experiments performed in triplicate. (B) HK-2, ACHN, Fa2 N-4, and HepG2 cells were treated with various concentrations of 2 as described in (A). After 48 h (Fa2 N-4 and HepG2) or 72 h (HK-2 and ACHN), cell extracts were prepared and immunoblotting analyses were performed to detect p21/Cip1, p16/INK4a, and FXR as described in Materials and methods.
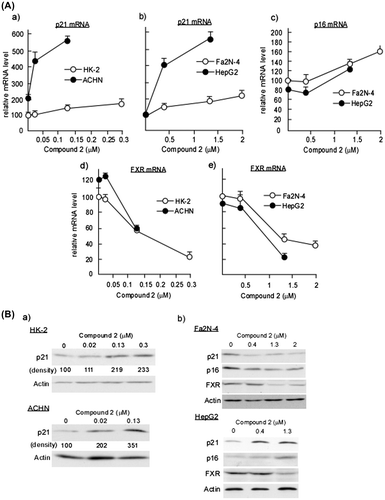
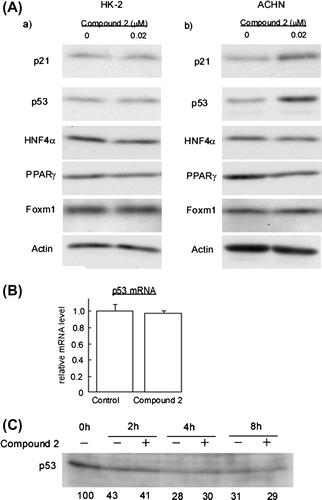
Fig. 6. Compound 2 upregulates p53 protein in ACHN Cells.
Notes: (A) HK-2 and ACHN cells were seeded at a density of 8.0 × 104 cells/35-mm dish. After 24 h, cells were treated with 0.02 μM for 72 h. Then cell extracts were prepared and immunoblotting analyses were performed to detect p21/Cip1, p53, HNF4α, PPARγ, and Foxm1. (B) ACHN cells were subjected to the 2 treatment as described in (A). Total RNA was extracted and quantification of p53 mRNA was performed as described in Materials and methods. Data are presented as the mean values ± SEM of three experiments performed in triplicate. (C) ACHN cells were seeded at a density of 2.0 × 105 cells/60-mm dish. After 24 h, 10 μg/ml cycloheximide was added to the culture and further incubated for the indicated times in the presence or absence of 0.02 μM 2. Then cell extracts were prepared and immunoblotting analyses were performed to detect.