Figures & data
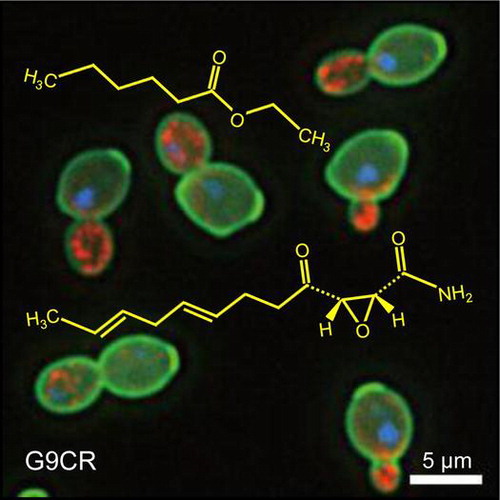
Fig. 1. Drug sensitivity of the yeast strains.
Notes: Drug sensitivity of laboratorial and industrial yeast strains. The laboratorial yeast strains, WT or Δbub1 (defective in SAC) and rad53 (defective in DIC), were used as the control strains, positive or negative, respectively. Drug sensitivities of a laboratorial yeast strain, Δrim15, and those of 4 industrial yeast strains, K7 (Brewing Society of Japan, Kyokai No.7), K1801 (Brewing Society of Japan, Kyokai No.1801), G9 (Niigata Prefectural Sake Research Institute), and the isolated G9CR, were examined on YPD plates containing cerulenin, benomyl, or HU at the indicated concentrations. The upper (WT, Δbub1, rad53, Δrim15, and K7) and lower panels (K1801, G9, and G9CR) are the left and right sides, respectively, on the same plate.
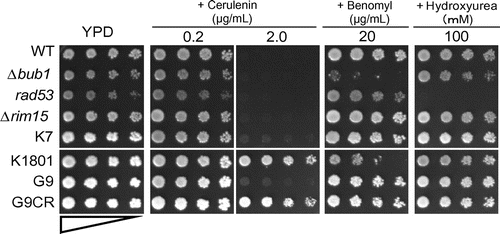
Fig. 2. Checkpoint integrity of the yeast strains.
Notes: (A, B) Cell viability of the indicated yeast strains in YPD liquid medium containing 15 μg/mL benomyl (A) or 200 mM HU (B) at the indicated times (h).
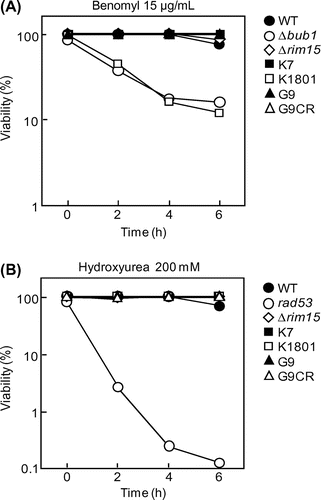
Fig. 3. Screening scheme for the spontaneous FAS2-G1250S yeast mutant with high productivity of ethyl caproate.
Notes: (A) Isolation of the FAS2-G1250S mutant with high productivity of ethyl caproate. At the first screening, cerulenin-resistant mutants were selected. At the second screening, the free fatty acid (FFA) high-producing mutants were selected by performing the FFA assay. At the third screening, the FAS2 mutation site in the selected mutants was examined by digestion with the restrictive enzyme Bfa I and DNA sequencing. (B) Confirmation of the FAS2 mutation site. The FAS2 gene in the mutant was amplified by PCR using 2 primers, P1 and P2. The amplified 503-bp fragment was digested by Bfa I. Putative cleavage DNA fragments of WT FAS2 gene and mutated FAS2-G1250S gene are 2 bands (430 and 73 bp) and 3 bands (205, 225, and 73 bp), respectively. (C) Cleavage pattern of the DNA sample after Bfa I-mediated digestion of the PCR-amplified fragment. Bfa I-digested (+) and non-digested DNA fragments (−) were loaded onto an agarose gel. M indicates the Gene Ladder Wide 1 (NIPPON GENE), which used as a size marker.
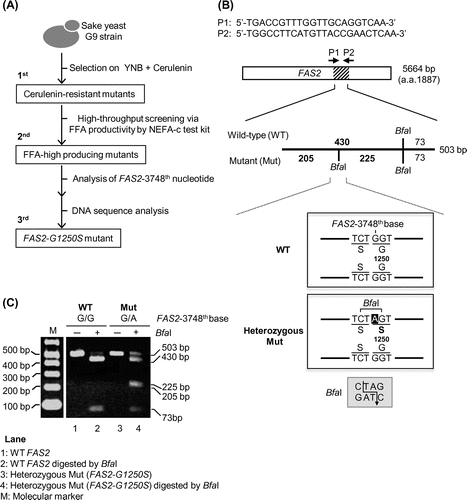
Fig. 4. Morphological phenotypic robustness of K1801 and G9CR.
Notes: (A) Morphology of K1801 and G9CR. The cells was examined after having been stained with FITC-ConA (green), Rh-ph (red), or DAPI (blue) to visualize the cell wall, actin, and nucleus, respectively. (B) Phenotypic potential of sake yeast was scored from each experiment data (n = 5). Asterisk indicates a significant difference (p < 0.05 by Student’s t-test) between the bracketed values. (C) Mean and noise score (normalized variance, [Citation19]) of the morphological parameter C117_A1B (cell size ratio) were obtained from K1801 and G9CR morphometric data.
![Fig. 4. Morphological phenotypic robustness of K1801 and G9CR.Notes: (A) Morphology of K1801 and G9CR. The cells was examined after having been stained with FITC-ConA (green), Rh-ph (red), or DAPI (blue) to visualize the cell wall, actin, and nucleus, respectively. (B) Phenotypic potential of sake yeast was scored from each experiment data (n = 5). Asterisk indicates a significant difference (p < 0.05 by Student’s t-test) between the bracketed values. (C) Mean and noise score (normalized variance, [Citation19]) of the morphological parameter C117_A1B (cell size ratio) were obtained from K1801 and G9CR morphometric data.](/cms/asset/26d1f6d6-440e-44d1-81c8-f42ac1d259e4/tbbb_a_1020756_f0004_oc.gif)
Table 1. Properties of sake made by G9, G9CR, and K1801 in small-scale sake brewing (1 kg of total rice).
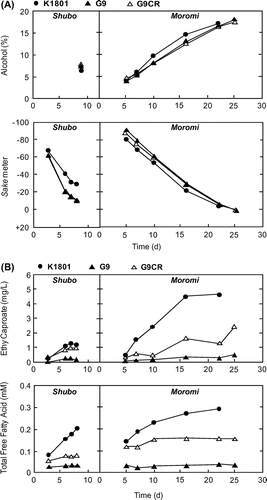
Fig. 5. Sake brewing test using K1801, G9, and G9CR.
Notes: (A) Fermentation profiles (upper, Alc; lower, Sm) in the sake mash (Shubo/pre-culture and Moromi/main culture) produced with K1801 (closed circle), G9 (closed triangle), or G9CR (open triangle). The values were measured by standard methods established by the National Tax Agency of Japan. (B) Contents of ethyl caproate (upper panel) and free fatty acids (lower panel) in the sake mash (Shubo/pre-culture and Moromi/main culture) produced with K1801, G9, or G9CR. Sake brewing test on an industrial scale was performed using 600 kg of the polished sake rice (Gohyakumangoku, polishing ratios of 50% for koji and 55% for rice added directly to the sake mash).