Figures & data
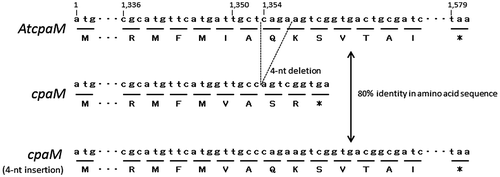
Fig. 1. Chemical structure of CPA,Citation1) 2-oxoCPACitation4) and speradine A (1-N-methyl-2-oxoCPA)Citation5) (A) and CPA biosynthesis gene cluster (cpa cluster) in Aspergillus oryzae NBRC 4177 (B).Citation4)
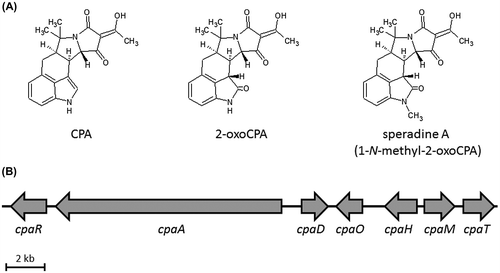
Fig. 2. Production of the CPA and the other derivatives in the transformants.
Notes: (A) Confirmation of transformation by PCR amplification of the introduced AtcpaM gene in the constructed strains, PAtcM-5 and -6. (B) UV chromatogram of 280 nm for the metabolite samples extracted from the culture broth of the transformants (PAtcM-5 and -6), A. oryzae NBRC 4177, and A. tamarii NBRC 4099. (C) The extracted ion chromatograms of m/z 337.155 (CPA), m/z 353.150 (2-oxoCPA), and m/z 367.165 (speradine A) in the metabolite samples of the transformants (PAtcM-5 and -6), A. oryzae NBRC 4177, and A. tamarii NBRC 4099. (D) MS/MS fragmentation analysis of the ions with of m/z 367.165 detected in the sample of A. oryzae transformant PAtcM-6, A. tamarii NBRC 4099 and the authentic speradine A. The collision energy was set to 24.5 eV.
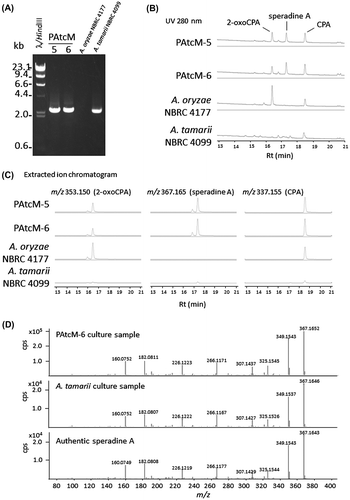
Fig. 3. Aligned sequences of nucleotides and amino acids of AtcpaM, cpaM, and cpaM with insertion of 4-nucleotide adjacent to the (corresponding) position where 4-nucleotide deletion occurs in cpaM.