Figures & data
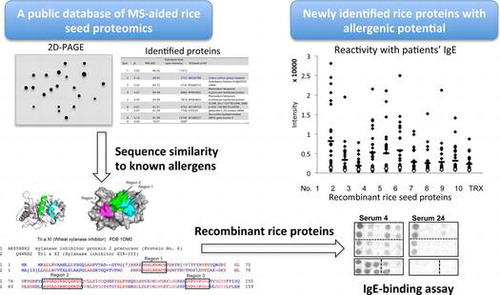
Table 1. Primer pairs for specific amplification of ORF for cDNAs encoding rice putative proteins. Recognition sites for restriction endonucleases are underlined.
Table 2. Some biochemical and structural properties of nine rice endosperm proteins with sequence similarity to known allergens and corresponding cDNA clones in the rice full-length cDNA database.
Fig. 1. Amino acid sequence alignment of putative rice proteins with sequence similarity to known allergens. (A) The sequences of three rice putative protein kinases (the rice proteins Nos. 1, 7, and 9 in Table ) were aligned with that of a pollen allergen from Russian thistle (Sal k 2). Regions are boxed at which six contiguous residues match between the rice protein and Sal k 2. (B) The sequence of rice putative germin-like protein (the rice protein No. 3 in Table ) was aligned with that of a fruit allergen (Cit l 1, germin-like protein) from lemon peel (flavedo). Regions are boxed at which eight contiguous residues or more match between the two. (C) The sequence of rice putative xylanase inhibitor (the rice protein No. 6 in Table ) was aligned with that of a food allergen (Tri a XI, xylanase inhibitor) from wheat seed. Regions are boxed at which nine contiguous residues or more match between the two. The crystal structure of wheat xylanase inhibitor (PDB: 1OM0) is shown as ribbon and surface models (Bottom). The matched contiguous residues in the three regions as well as the disulfide bridge between Cys55 and Cys96 are colored, respectively.
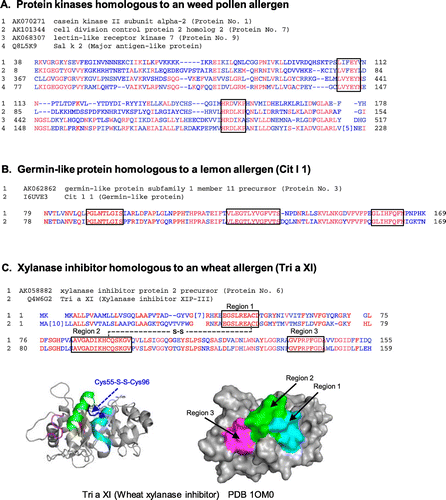
Fig. 2. Expression and affinity purification of recombinant rice proteins and total-IgE concentration of patients’ serum specimens. (A) The putative rice seed proteins encoded by the nine full-length cDNA clones (GeneBank ID: AK070271, AK119653, AK062862, AK073487, AK242325, AK058882, AK101344, AK242260, and AK068307) were expressed as His-tagged TRX-fusion proteins by an E. coli expression system and purified using Ni-affinity chromatography. Total proteins of transformed E. coli lysate (T) and affinity-purified proteins (P) were analyzed by SDS-PAGE (12.5% acryl amide gel stained with CBB R-250). The protein number (1–9) in this study is shown above each strip of the PAGE gel. The bands of His-tagged TRX-fusion proteins are indicated with black arrowhead. (B Total-IgE concentration of the patients’ serum specimens (rice-positive, Nos. 1 to 26; rice-negative, Nos. 27 and 28) was determined by two-site ELISA using two kinds of anti-human IgE as described in Materials and Methods. The total-IgE concentration is expressed as international units (IU) per mL.
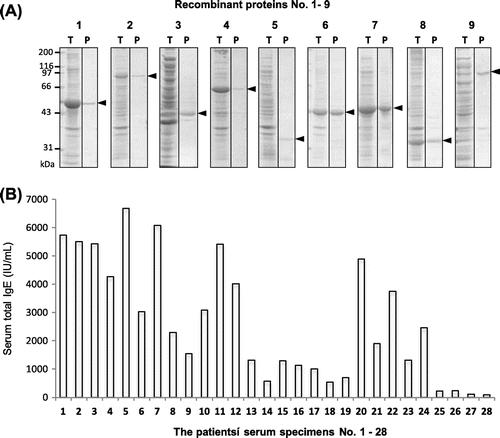
Fig. 3. IgE-binding to recombinant rice proteins with sequence similarity to known allergens.
Fig. 4. Semi-quantitative analysis of IgE-binding to recombinant rice proteins as estimated by immuno-dot blotting. (A) Signal intensity of IgE-binding to each protein dot for the 28 serum specimens was semi-quantified using Image-J and plotted on the nine recombinant rice proteins and the control proteins, LTP, TRX, rice seed proteins, and egg albumin (EA). The mean value of the 26 positive sera is also shown as a horizontal bar. (B) TRX-rice protein fusion/TRX ratio in the IgE-binding intensity is compiled as a matrix of the nine rice recombinant proteins and the 28 serum specimens including 2 negative controls. The relative values above 2.0 and 5.0, which were regarded, respectively, as positive and strong positive in the IgE-binding, were highlighted with gray and black backgrounds, respectively.
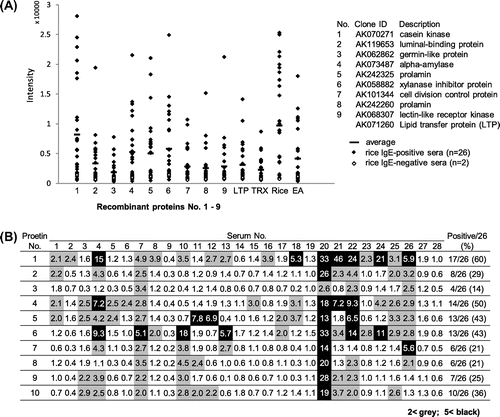
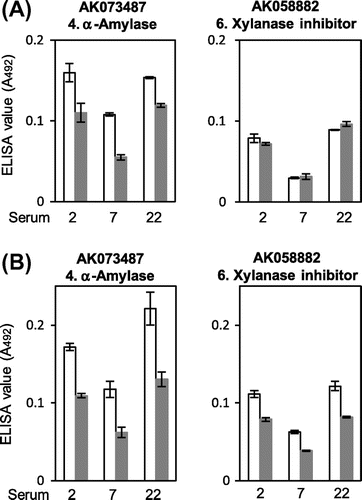
Fig. 5. Effect of heat-induced denaturation on the IgE-binding to putative rice α-amylase and xylanase inhibitor. The recombinant proteins of putative rice α-amylase (AK073487) and xylanase inhibitor (AK058882) were heat denatured by boiling in the absence (A) and presence (B) of 2-mercaptoethanol and then subjected to the quantitative IgE-binding analysis using ELISA on the three serum specimens, which were IgE-binding positive and available relatively in large amount. The ELISA values for native (white bars) and heat-denatured (gray bars) proteins are shown as mean ± SE of three determinations.