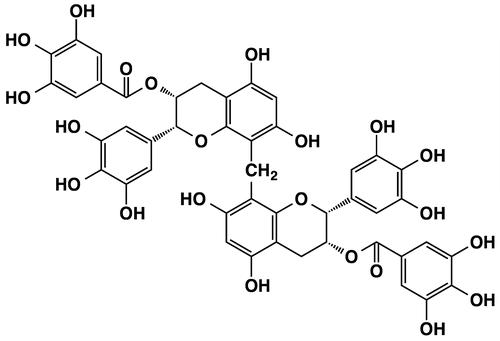
Figures & data
Fig. 2. DPPH radical scavenging capacity of OFA.
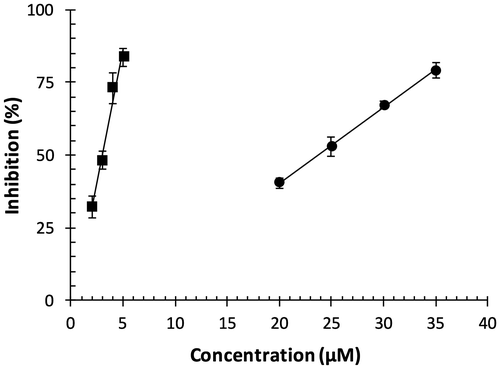
Fig. 3. Effect of OFA on peroxyl radical-mediated lipid peroxidation of LDL.
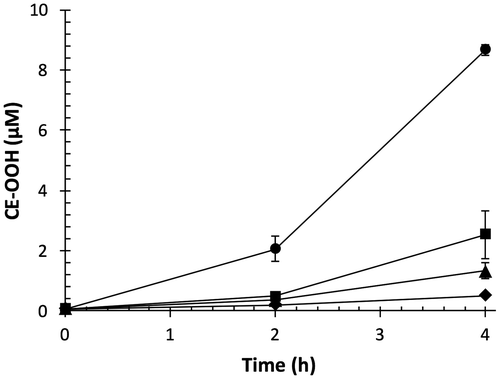
Fig. 4. Effect of OFA on transition metal ion-catalyzed lipid peroxidation of LDL.
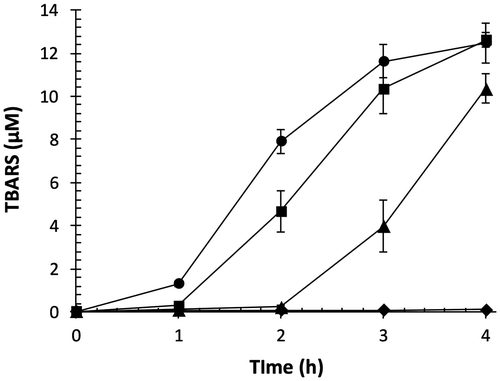
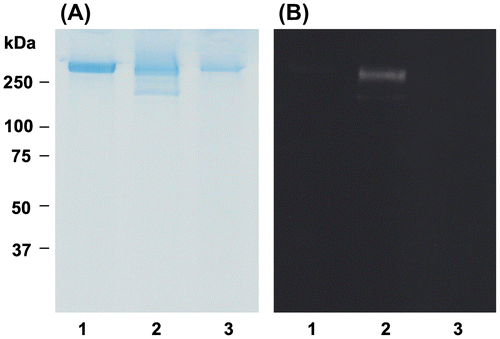
Fig. 5. Effect of OFA on apo B-100 modification in oxLDL.
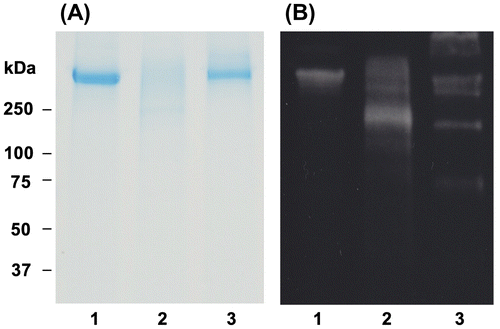
Fig. 6. Effect of OFA on protein carbonyl formation in oxLDL.
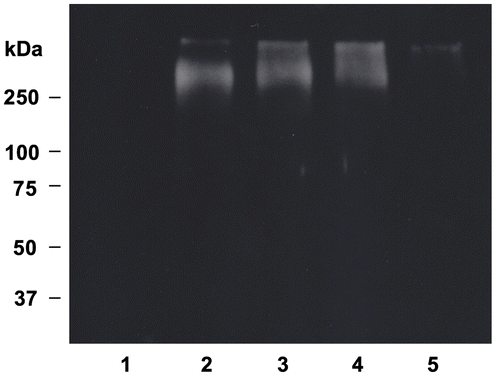
Fig. 7. Effect of OFA on heparin-binding activity of apo B-100 in oxLDL.
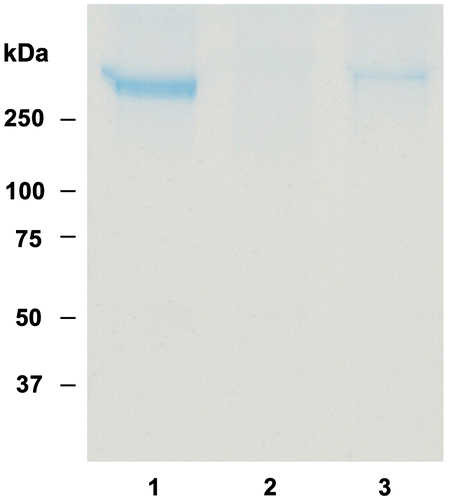
Fig. 8. Effect of OFA on peroxynitrite-mediated lipid peroxidation of LDL.
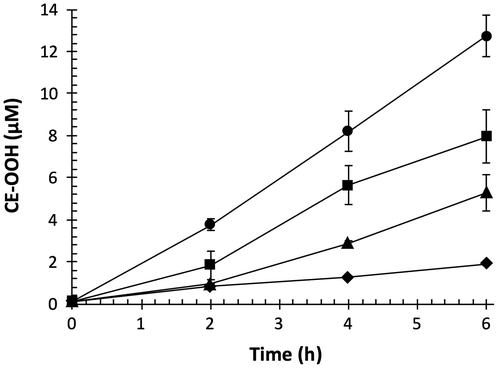
Fig. 9. Effect of OFA on nitrotyrosine formation in oxLDL.