Figures & data
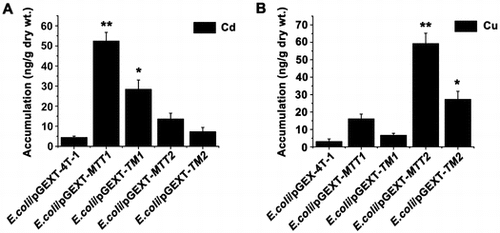
Table 1. PCR primers used in this study.
Figure 1. Amino acid sequences of MTT1 and MTT2 from T. thermophila. The C3X6C2, C2X6/8CXCX2CXC2X1/2, and CXC represent different motifs. TM1 and TM2 indicate the truncated MTT1 and MTT2, respectively.
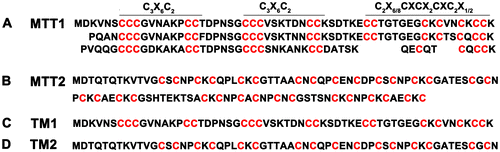
Figure 2. Expression and purification of GST, GST-MTT1, GST-MTT2, GST-TM1, and GST-TM2. (A) SDS-PAGE (12.5%) analysis of recombinant proteins. M: protein molecular weight markers; lane 1: total cell lysate of E. coli/pGEX-4T-1; lane 2: total cell lysate of E. coli/pGEX-MTT1; lane 3: total cell lysate of E. coli /pGEX-MTT2; lane 4: total cell lysate of E. coli/pGEX-TM1; lane 5: total cell lysate of E. coli /pGEX-TM2. (B) SDS-PAGE (12.5%) analysis of purified proteins. M: protein molecular weight markers; lane 1: GST; lane 2: GST-MTT1; lane 3: GST-MTT2; lane 4: GST-TM1; lane 5: GST-TM2. The proteins were purified using glutathione resins and Superdex 75 gel filtration chromatography.
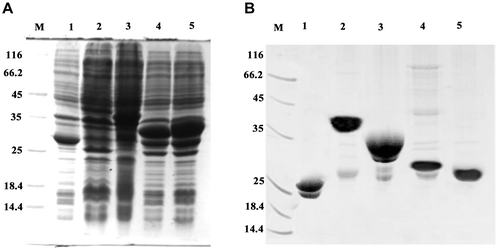
Figure 3. Cd and Cu dose–response curve of recombinant MT E. coli. Recombinant E. coli/pGEX-MTT1, E. coli/pGEX-MTT2, E. coli/pGEX-TM1, and E. coli/pGEX-TM2 were exposed to CdCl2 (A) and CuSO4 (B). Cell concentration was measured at 600 nm using a spectrophotometer. All data are expressed as mean ± SD (n = 3).
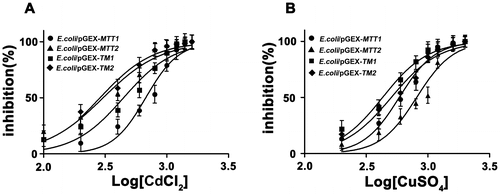
Figure 4. Proliferation of recombinant MT in E. coli. Recombinant E. coli/pGEX-MTT1, E. coli/pGEX-MTT2, E. coli/pGEX-TM1, and E. coli/pGEX-TM2 were exposed to 287.1 μM CdCl2 (A) and 429.5 μM CuSO4 (B). E. coli/pGEX-4T-1 as a control, cell concentration was measured at 600 nm using a spectrophotometer. All data are expressed as mean ± SD (n = 3).
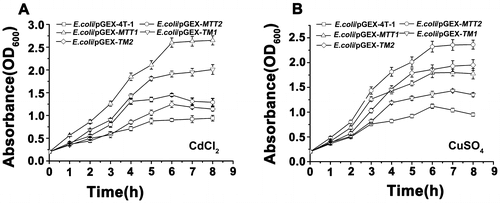
Figure 5. Accumulation of metal ion Cd2+ and Cu+ in MT transformed cells. Recombinant E. coli/pGEX-MTT1, E. coli/pGEX-MTT2, E. coli/pGEX-TM1, and E. coli/pGEX-TM2 were exposed to 287.1 μM CdCl2 (A) and 429.5 μM CuSO4 (B). E. coli/pGEX-4T-1 as a control. The metal ion was analyzed using an atomic absorption spectrometer. All data are expressed as mean ± SD (n = 3), *p < 0.05, **p < 0.001.
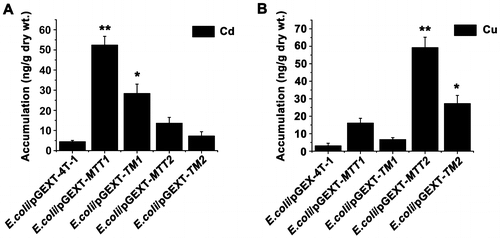
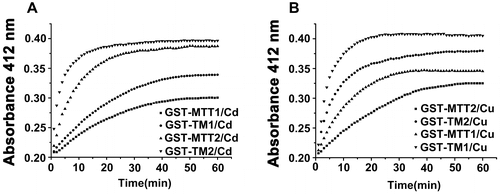
Figure 6. Reaction profiles of metal-binding complexs with DTNB. Each metal-bound protein sample at 1.5 nmol was dissolved in 300 μL of 10 mM PBS (pH 8.0), and reaction was started by adding 75 nmol of DTNB to solution. Absorbance at 412 nm at 25 °C was recorded for 60 min. A: Cd-bound protein sample; B: Cu-bound protein sample. Each value was the mean of two replicates.