Figures & data
Table 1. Primers and probes used in this study for RT-PCR analysis.
Figure 1. Common laboratory findings and clinical disorders observed in SLE patients participated in this study.
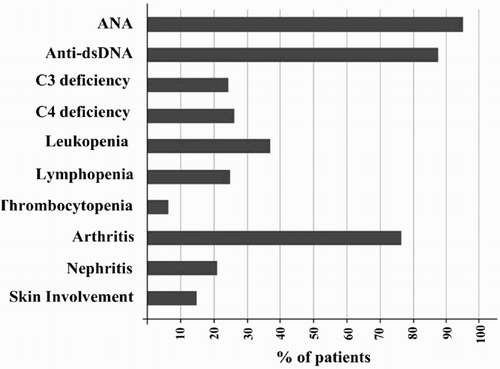
Figure 2. Representative flow cytometric analysis of CD4+CD25high Foxp+ regulatory T-cells among vitamin D treated (D+) and untreated (D-) PBMCs of SLE patients. CD4+ cells were gated and percentage of CD25high Foxp3+ cells was analyzed.
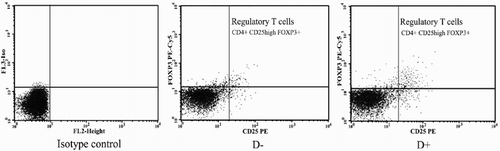
Figure 3. RT-PCR analysis using comparative CT method to quantify gene expression levels of Foxp3, TGFβ, and IL6. Data expressed as mean transcript expression fold-change over untreated samples (controls) normalized to GAPDH. Quantitative RT-PCR analysis of the vitamin D-treated cells revealed that transcript expression of Foxp3 (1.41 ± 0.30), TGFβ (1.38 ± 0.24), and IL6 (0.50 ± 0.09) underwent fold-changes relative to the levels in untreated cells. This translated to up-regulated expressions of Foxp3 (41%), TGFβ (38%) and down-regulated expressions of IL6 (50%) due to culture in the presence of 50 nM vitamin D. Values are shown as mean ± SD.
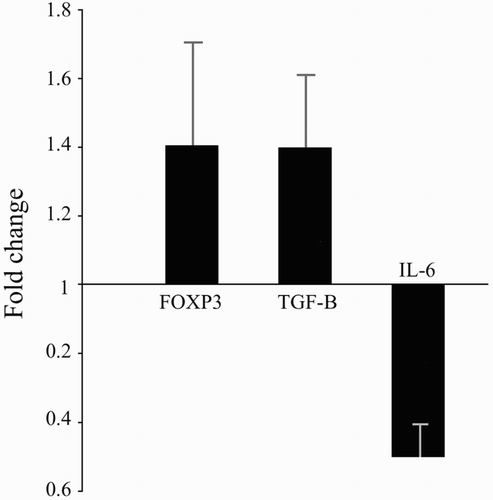
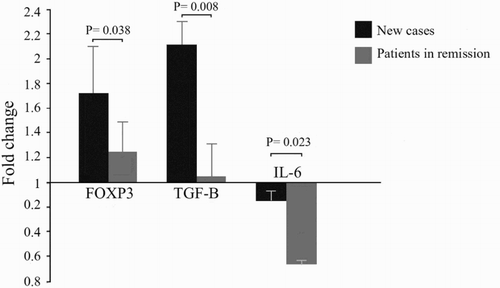
Figure 4. Comparison of gene expression results between new cases and in remission SLE patients. Vitamin D treatment up-regulated the expression levels of Foxp3 (1.72 ± 0.36 vs. 1.23 ± 0.26) (p = .038), and TGFβ (2.11 ± 0.21 vs. 1.04 ± 0.28) (p = .008) in new cases more significantly compared to SLE patients who were in remission. Vitamin D treatment in SLE patients who were in remission down-regulated the expression of IL6 more significantly compared to new cases (0.35 ± 0.05 vs. 0.85 ± 0.12) (p = .023).