Figures & data
Table 1. Summary of proteolytic conditions.
Figure 1. Proteolysis of prolamins detected by SDS-PAGE (a) and Western blot (b) after 60 min of hydrolysis using protease from Bacillus licheniformis: CW1 – common wheat, cultivar Saxana, spring form; CW2 – common wheat, cultivar Blava, winter form; DW – durum wheat, cultivar Soldur; SW – spelt wheat, cultivar Rubiota; R – rye, cultivar Dankowskie Nowe; T – triticale, cultivar Wanad; B – barley, cultivar Ludan; O – oat, cultivar Detvan; lane 1 – prolamins treated using protease; lane 2 – untreated prolamins; M – molecular marker; arrows indicate fragments after proteolysis.
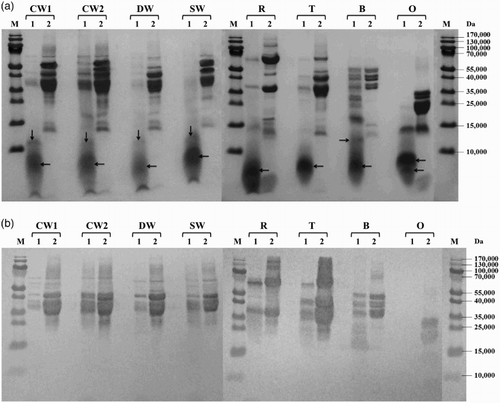
Figure 2. Proteolysis of prolamins detected by SDS-PAGE (a) and Western blot (b) after 60 min of hydrolysis using protease from B. stearothermophilus: CW1 – common wheat, cultivar Saxana, spring form; CW2 – common wheat, cultivar Blava, winter form; DW – durum wheat, cultivar Soldur; SW – spelt wheat, cultivar Rubiota; R – rye, cultivar Dankowskie Nowe; T – triticale, cultivar Wanad; B – barley, cultivar Ludan; O – oat, cultivar Detvan; lane 1 – prolamins treated using protease; lane 2 – untreated prolamins; M – molecular marker; arrows indicate fragments after proteolysis.
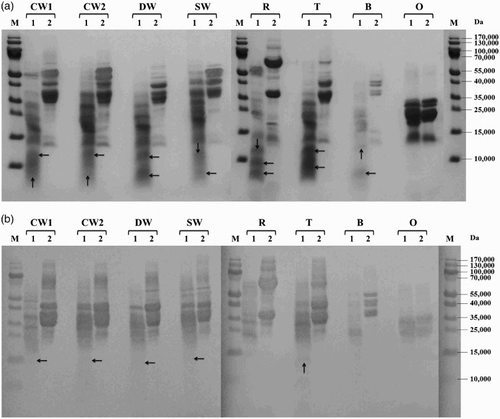
Figure 3. Proteolysis of prolamins detected by SDS-PAGE (a) and Western blot (b) after 60 min of hydrolysis using protease from Bacillus thermoproteolyticus: CW1 – common wheat, cultivar Saxana, spring form; CW2 – common wheat, cultivar Blava, winter form; DW – durum wheat, cultivar Soldur; SW – spelt wheat, cultivar Rubiota; R – rye, cultivar Dankowskie Nowe; T – triticale, cultivar Wanad; B – barley, cultivar Ludan; O – oat, cultivar Detvan; lane 1 – prolamins treated using protease; lane 2 – untreated prolamins; M – molecular marker; arrows indicate fragments after proteolysis.
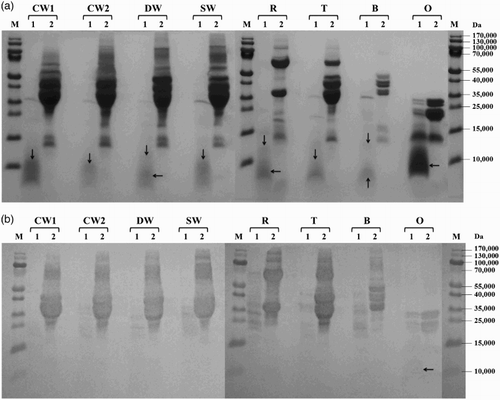
Figure 4. Proteolysis of prolamins detected by SDS-PAGE (a) and Western blot (b) after 60 min of hydrolysis using protease from Streptomyces griseus: CW1 – common wheat, cultivar Saxana, spring form; CW2 – common wheat, cultivar Blava, winter form; DW – durum wheat, cultivar Soldur; SW – spelt wheat, cultivar Rubiota; R – rye, cultivar Dankowskie Nowe; T – triticale, cultivar Wanad; B – barley, cultivar Ludan; O – oat, cultivar Detvan; lane 1 – prolamins treated using protease; lane 2 – untreated prolamins; M – molecular marker; arrows indicate fragments after proteolysis.
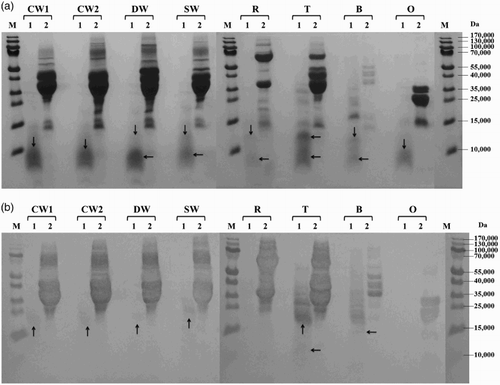
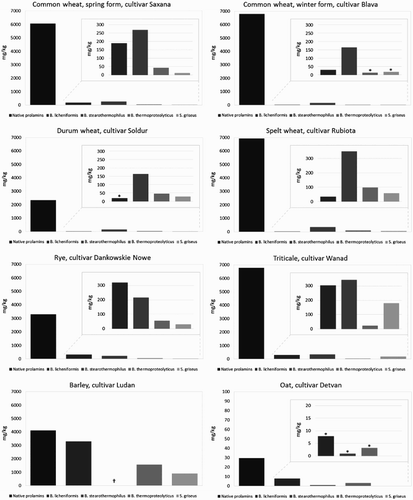
Figure 5. Proteolysis of prolamins detected by R5 ELISA after 60 min of hydrolysis using different microbial proteases; *ppm values lower than 20 mg/kg (Codex Alimentarius limit); †inconclusive results; ‡<LOD (limit of detection).