Figures & data
Table 1. Phylogenetic attribution of isolates from cultures of Tetraselmis suecica F&M-M33, based on 16S rDNA sequencing
Table 2. Identification of bacteria isolates producing at least a microalgal growth promoting compounds (MGP compounds): indole-3-acetic acid (IAA) and/or siderophores
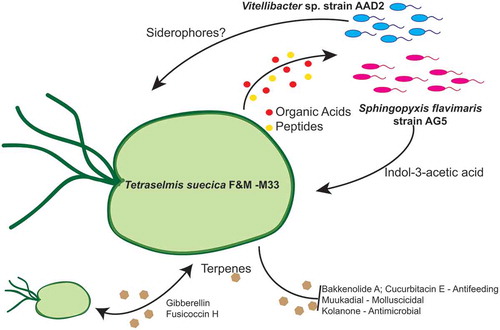
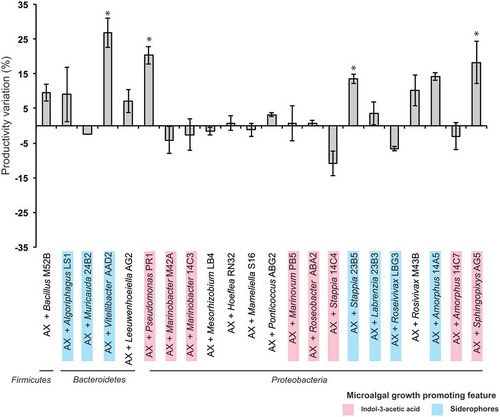
Fig. 1. Tetraselmis suecica F&M-M33 biomass productivity. Co-cultures were obtained using an axenic culture of T. suecica F&M-M33 (AX) inoculated with a selected bacterial strain (AX + bacterial strain name). Strains with microalgal growth-promoting features are highlighted in pink (indol-3-acetic acid producers) or blue (siderophore producers). Co-cultures and axenic cultures were grown for 10 days at 27°C and under continuous illumination of 90 µmol photons m−2 s−1 and shaking (85 rpm). Bars show the relative percentage difference of productivity between co-cultures and axenic culture. * indicates statistically significant differences (ANOVA, p < 0.05)
Figs 2–3. Venn diagrams showing unique entities, and entities shared by all or by two culture supernatants of axenic Tetraselmis suecica F&M-M33 (AX) and co-cultures of the axenic T. suecica F&M-M33 with either Sphingopyxis flavimaris strain AG5 (AX+AG5) or Vitellibacter strain AAD2 (AX+AAD2). Fig. 2. Positive ionization mode. Fig. 3. Negative ionization mode
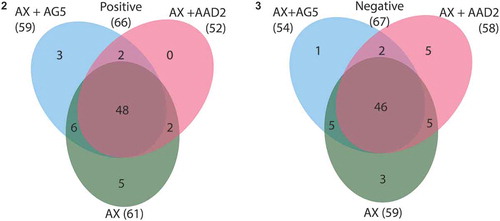
Table 3. Entities (small chemical molecules, e.g. metabolites) retrieved in the supernatant of Tetraselmis suecica F&M-M33 axenic culture (AX) only and entities exclusively present in the supernatants of T. suecica F&M-M33 co-cultured with Vitellibacter strain AAD2 (AX+AAD2) or with Sphingopyxis flavimaris strain AG5 (AX+AG5). Data are referring to the analysis performed in positive (pos) and negative (neg) ionization modes