Figures & data
Fig. 1. Schematic representation of the experimental design, depicting the division of Drosophila melanogaster individuals into two major groups: one unchallenged and another streptonigrin-challenged (S). Each of the previous groups was divided into two subgroups, corresponding to wild-harvested and aquacultured Ulva rigida (U) trials (depicted in the groups codes 1 and 2, respectively). For each subgroup, two levels of supplementation (2.5 and 5%; represented in the groups codes by the number preceding the letter U) and a control group (with no alga supplementation; C or S) were tested
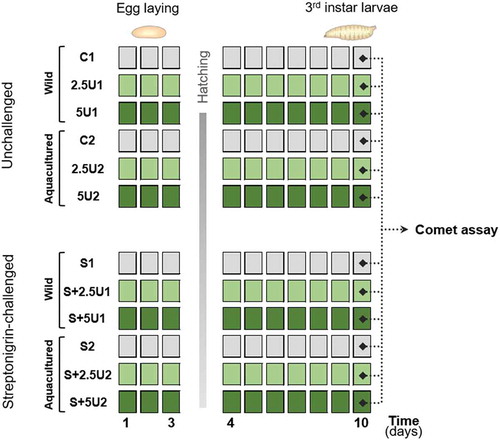
Table 1. Compounds identified from the hexane extracts of wild-harvested and aquacultured Ulva rigida
Table 2. Compounds identified from ethanolic extracts of wild-harvested and aquacultured Ulva rigida, and their molecular ions species and fragments (m/z) data
Fig. 2. Mean values of DNA damage (GDI) in Drosophila melanogaster neuroblasts after a supplementation with Ulva rigida (2.5 and 5%; number preceding the letter U) from wild-harvesting (U1) and aquaculture (U2) origins. C and S represent the unsupplemented unchallenged and streptonigrin-challenged groups, respectively. Bars represent the standard error. Statistically significant differences between groups submitted to the supplementation with U. rigida of the same origin, under the same genotoxic challenge are marked by different letters; differences between U1 and U2 groups under the same genotoxic challenge and level of supplementation are marked by black diamond (♦); differences between groups unchallenged and streptonigrin-challenged, under the same level of supplementation, from the same origin are marked by asterisk (*)
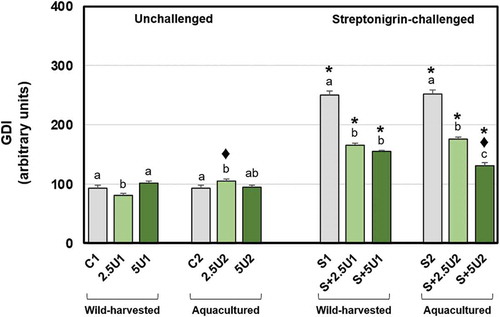
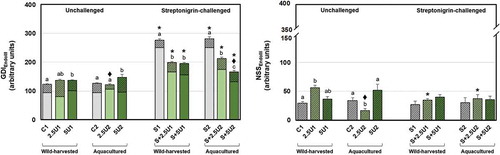
Fig. 3-4. Mean values of oxidative purine DNA damage in Drosophila melanogaster neuroblasts after a supplementation with Ulva rigida (2.5 and 5%; number preceding the letter U) from wild-harvesting (U1) and aquaculture (U2) origins. C and S represent the unsupplemented unchallenged and streptonigrin-challenged groups, respectively. (Fig. 3) overall damage (GDIFPG) and partial scores, i.e. GDI plus additional DNA breaks corresponding to net FPG-sensitive sites (NSSFPG; dark grey diagonal stripes); (Fig. 4) NSSFPG alone. Bars represent the standard error. Statistically significant differences between groups submitted to the supplementation with U. rigida of the same origin, under the same genotoxic challenge are marked by different letters; differences between U1 and U2 groups under the same genotoxic challenge and level of supplementation are marked by black diamond (♦); differences between groups unchallenged and streptonigrin-challenged, under the same level of supplementation, from the same origin are marked by asterisk (*)
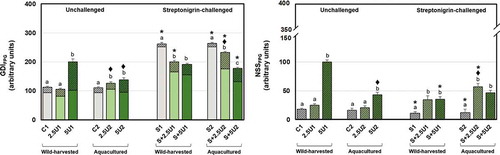
Fig. 5-6. Mean values of oxidative pyrimidine DNA damage in Drosophila melanogaster neuroblasts after a supplementation with Ulva rigida (2.5 and 5%; number preceding the letter U) from wild-harvesting (U1) and aquaculture (U2) origins. C and S represent the unsupplemented unchallenged and streptonigrin-challenged groups, respectively. (Fig. 5) overall damage (GDIEndoIII) and partial scores, i.e. GDI plus additional DNA breaks corresponding to net EndoIII-sensitive sites (NSSEndoIII; dark grey diagonal stripes); (Fig. 6) NSSEndoIII alone. Bars represent the standard error. Statistically significant differences between groups submitted to the supplementation with U. rigida of the same origin, under the same genotoxic challenge are marked by different letters; differences between U1 and U2 groups under the same genotoxic challenge and level of supplementation are marked by black diamond (♦); differences between groups unchallenged and streptonigrin-challenged, under the same level of supplementation, from the same origin are marked by asterisk (*)