Figures & data
Figure 1. Illustration of KIF2C’s phosphorylation sites, domains, interaction partners and molecular functions. (A): KIF2C’s domains, functions and phosphorylation sites are depicted. The phosphorylation sites are marked as green for Aurora A, blue for Aurora B, light blue for Cdk1, red for Plk1, purple for PAK1 and black for a phosphorylation by an unknown kinase. (B): schematic illustration of KIF2C/MCAK’s molecular involvement in regulating chromosome congression/segregation, FA turnover, ciliogenesis and DSB repair. The figure was modified and updated from [Citation17]. Abbreviations: Cdk1: Cyclin-dependent kinase 1; Plk1: Polo-like kinase 1; PAK1: p21-activated kinase; EB1: end-binding protein 1; FA: focal adhesion; DSB: double-strand break; NT: NH2-terminus; CT: carboxy-terminus; NHEJ: non-homologous end joining; HR: homologous recombination.
![Figure 1. Illustration of KIF2C’s phosphorylation sites, domains, interaction partners and molecular functions. (A): KIF2C’s domains, functions and phosphorylation sites are depicted. The phosphorylation sites are marked as green for Aurora A, blue for Aurora B, light blue for Cdk1, red for Plk1, purple for PAK1 and black for a phosphorylation by an unknown kinase. (B): schematic illustration of KIF2C/MCAK’s molecular involvement in regulating chromosome congression/segregation, FA turnover, ciliogenesis and DSB repair. The figure was modified and updated from [Citation17]. Abbreviations: Cdk1: Cyclin-dependent kinase 1; Plk1: Polo-like kinase 1; PAK1: p21-activated kinase; EB1: end-binding protein 1; FA: focal adhesion; DSB: double-strand break; NT: NH2-terminus; CT: carboxy-terminus; NHEJ: non-homologous end joining; HR: homologous recombination.](/cms/asset/a916fb54-b059-4b94-93e7-4372f196e662/ilab_a_2309933_f0001_c.jpg)
Table 1. Correlation between KIF2C/MCAK expression and the clinicopathologic features of various cancer entities.
Table 2. Molecular mechanisms involved in KIF2Cs oncogenic potential in multiple cancer entities/cell lines.
Figure 2. The model depicts how deregulated KIF2C/MCAK is associated with some of the fourteen known and discussed hallmarks of cancer including acquired capabilities and enabling characteristics [Citation84]. These hallmarks encompass the following terms: deregulated cellular metabolism, sustaining proliferative signaling, resisting cell death, evading growth suppressors, polymorphic microbiomes, enabling replicative immortality, avoiding immune destruction, nonmutational epigenetic reprogramming, unlocking phenotypic plasticity, activating invasion and metastasis, tumor-promoting inflammation, cellular senescence, inducing or accessing vasculature and genome instability and mutation. Deregulated KIF2C may be associated with at least five of these hallmarks by promoting migration and invasion, disrupting MT dynamics, impairing the immune response, inducing senescence and increasing therapy resistance to MT interfering agents.
![Figure 2. The model depicts how deregulated KIF2C/MCAK is associated with some of the fourteen known and discussed hallmarks of cancer including acquired capabilities and enabling characteristics [Citation84]. These hallmarks encompass the following terms: deregulated cellular metabolism, sustaining proliferative signaling, resisting cell death, evading growth suppressors, polymorphic microbiomes, enabling replicative immortality, avoiding immune destruction, nonmutational epigenetic reprogramming, unlocking phenotypic plasticity, activating invasion and metastasis, tumor-promoting inflammation, cellular senescence, inducing or accessing vasculature and genome instability and mutation. Deregulated KIF2C may be associated with at least five of these hallmarks by promoting migration and invasion, disrupting MT dynamics, impairing the immune response, inducing senescence and increasing therapy resistance to MT interfering agents.](/cms/asset/a3046ef3-9adb-47a2-a977-bc00b8d3a426/ilab_a_2309933_f0002_c.jpg)
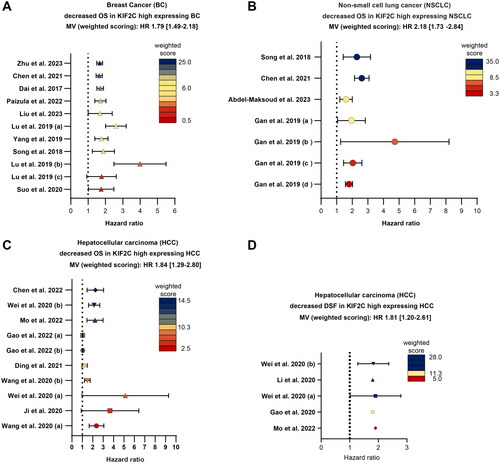
Figure 3. High expression of KIF2C is associated with reduced OS and DSF in BC, HCC and NSCLC. (A–C) The meta-analyses concerning KIF2C/MCAK’s importance as a prognostic marker is depicted as forest plots for OS of BC, NSCLC and HCC patients (A–C). (D) Meta-quantification of KIF2C/MCAK involvement in DSF in HCC patients. All individual studies were scored based on their patient cohort. The weighted score (ws) is visualized in a heatmap, dark blue (high ws), yellow (intermediate ws) and red (low ws). The overall mean value (MV) extracted from all studies was calculated by integrating the weighted score. Abbreviations: OS: overall survival; DFS: disease-free survival.
Data availability statement
All data generated or analyzed during this study are included in this article and its supplementary information.
