Figures & data
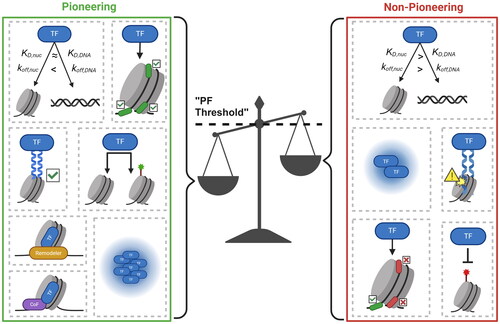
Figure 1. Some evidence suggests that PFs are a unique subset of TFs. (a) The apparent binding affinity of PFs for target sites on nucleosomes (KD,nuc) is similar to that of free DNA (KD,DNA). (b) some PFs have a stronger binding affinity for nucleosomal sites due to a dissociation rate compensation mechanism, where the nucleosomal off-rate (koff,nuc) is lower than the off-rate from free DNA (koff,DNA). (c) many PFs possess unique structural features in their DBDs that can better accommodate nucleosome substrates. (d) PFs are able to target internal nucleosomal motifs, like those at the dyad, while non-PFs primarily target motifs on free DNA or at the nucleosome entry-exit site. (e) PFs can target nucleosomal sites irrespective of the underlying chromatin landscape, whereas the binding of non-PFs tends to be inhibited at unmodified and repressive (red) sites but permitted at activating (green) sites. (f) PFs are the first TFs to bind to cis-regulatory elements that initiate gene regulatory events. Non-PFs tend to bind more downstream. (g) PFs can remain bound to chromatin throughout mitosis, whereas non-PFs dissociate from mitotic chromosomes.
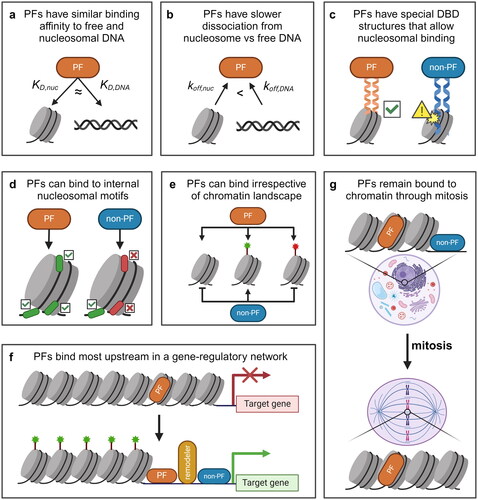
Figure 2. Some evidence suggests that PFs are not special and all/many TFs can function as PFs. (a) TF concentration correlates positively with nucleosomal targeting and opening. (b) Some PFs do not exhibit the dissociation rate compensation and have a higher nucleosomal off-rate (koff,nuc) than the off-rate from free DNA (koff,DNA). (c) many PFs have a stronger preference for binding to free DNA or at the nucleosome entry-exit site than to internal nucleosomal motifs. (d) Some evidence suggests that PFs are able to target unmarked and activating (green) sites but unable to target repressive (red) sites. (e) Some PFs are not retained on mitotic chromatin, whilst some TFs not characterized as PFs have been shown to remain bound to mitotic chromatin. (f) Some PFs rely on co-binding with other TFs (green or purple) to direct context-specific binding. (g) Some PFs rely on ATP-dependent chromatin remodelers for binding, despite the definition of PFs as being the most upstream factors in a gene regulatory pathway.
Table 1. Standard assay for PF classification.
Figure 3. Pioneer binding and activity is determined by a combination of intrinsic protein characteristics and context-dependent factors. A TF will bind to and potentially open a nucleosomal site given sufficient “pioneering” features. Pioneering features can be protein-intrinsic (having a low KD,nuc and koff,nuc, targeting internal nucleosomal motifs, possessing a nucleosome-compatible DBD, engaging in physical histone contacts, binding to unmarked/repressive chromatin, and recruiting chromatin remodelers) or context-specific (high TF concentration and availability of cofactors).