Figures & data
FIGURE 1 Total protein contents in a: whole fruit; b: flesh; and c: peel of nine pear cultivars in mg/g dry weight. Value are means ± SD (n = 3). Different letters (a–h) of each indicate significant difference within column at p < 0.05 by DMR test. The cultivars used were Chuwhang (CW), Shinwha (SW), Whasan (WS), Gamcheon (GC), Manpung (MP), Mansu (MS), Wonwhang (WW), Nikita (NK), and Hanareum (HA).
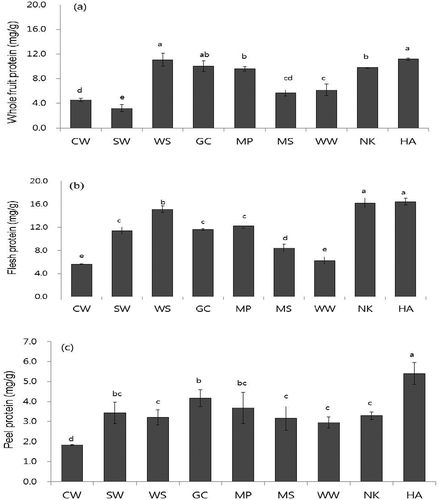
FIGURE 2 SDS-PAGE; a: analysis; and b: native-PAGE zymography of nine pear cultivars. M: molecular weight marker. Arrow indicates the protease bands of pear cultivars with the expected molecular weights of 36 and 38 kDa. The cultivars used were Chuwhang (CW), Shinwha (SW), Whasan (WS), Gamcheon (GC), Manpung (MP), Mansu (MS), Wonwhang (WW), Nikita (NK), and Hanareum (HA).
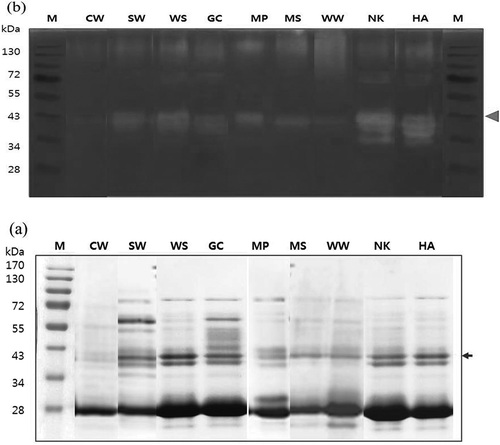
TABLE 1 Esterase activity of nine pear cultivars
TABLE 2 Caseinolytic activity of nine pear cultivars
TABLE 3 Collagenase activity of nine pear cultivars
FIGURE 3 Identification of pear protease by in-gel trypsinization and MALDI-TOF analysis. a: Coomassie-stained gel of Nikita pear. Arrow indicates the bands excised for in gel digestion. b: Peptide mapping spectrum. Numbered peaks are matched mass peaks with expected sequence of cysteine protease. c: Peptide mass peak list. d: Sequence coverage through the peptide finger printing analysis by MALDI-TOF.
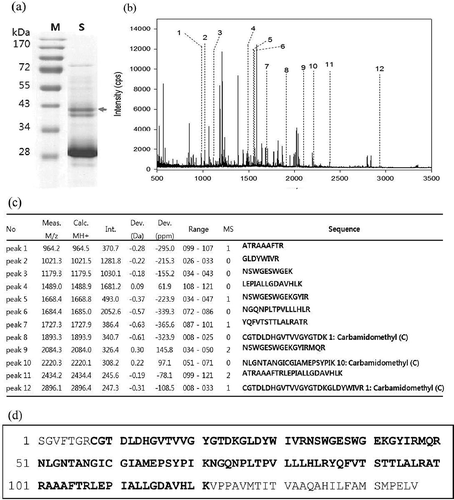
TABLE 4 Cysteine protease contents of nine pear cultivars
