Figures & data
Figure 1. Effect of short- and medium-chain FAs on the CI (circle: C10; triangle: C8; square: C6; diamond: C2). The values are expressed as mean ± SD (n = 3). Values with the same letter at each addition are not significantly different at p < 0.05.
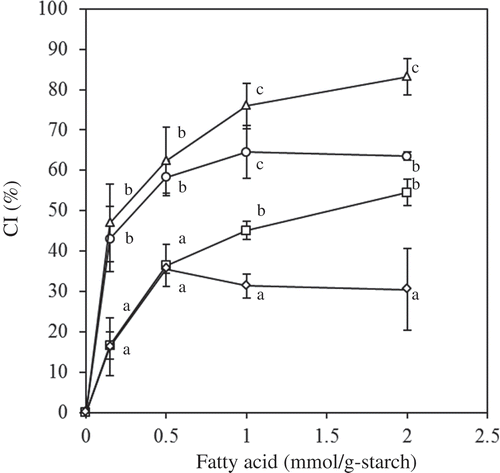
Figure 2. Effect of chain length in FAs on the CI observed at the maximum concentration tested (2 mmol/g starch). The values are expressed as mean ± SD (n = 3).
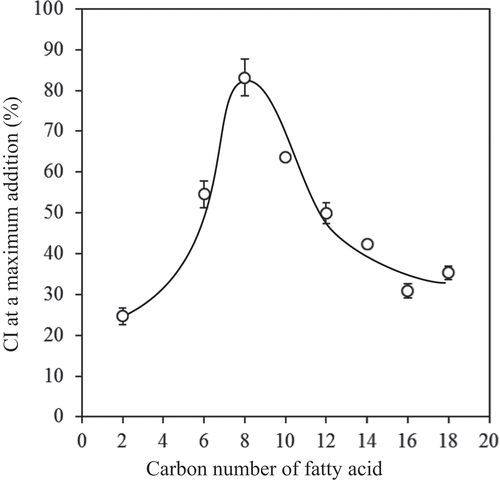
Figure 3. Effect of MGs on the CI (circle: MG-C12; triangle: MG-C14; square: MG-C16; open diamond: MG-C18). The values are expressed as mean ± SD (n = 3). Values with the same letter at each addition are not significantly different at p < 0.05.
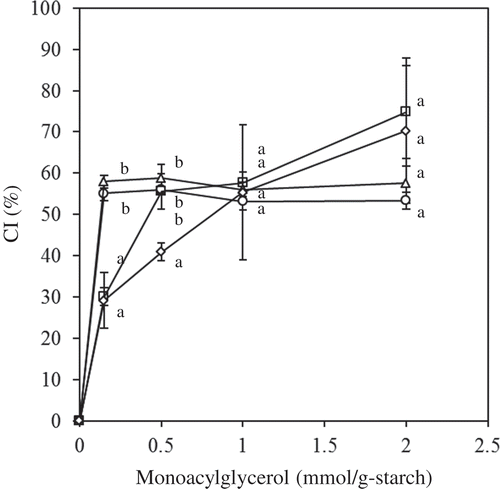
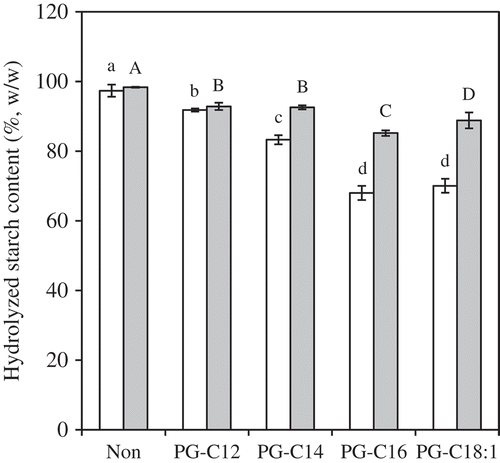
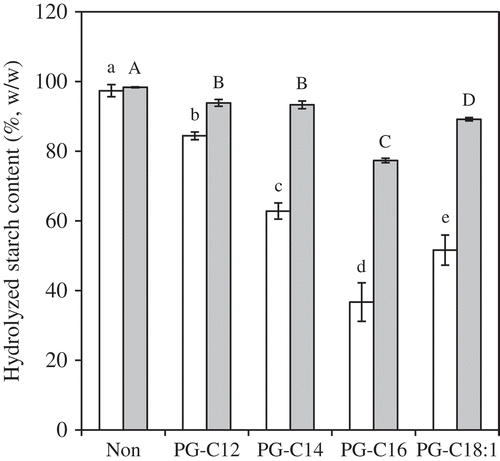
Figure 4. HS20 (white) and HS120 (gray) for MG-samples (0.15 mmol/g starch). The values are expressed as mean ± SD (n = 3). Values with the same letter at each hydrolyzing condition are not significantly different at p < 0.05.
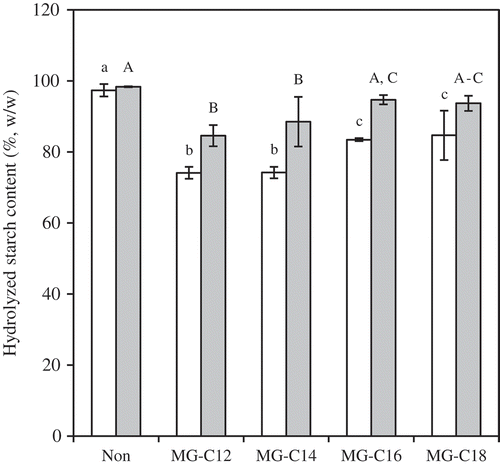
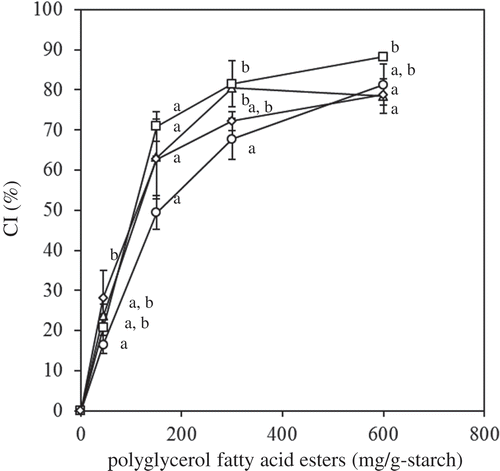
Figure 5. Effect of PGs on the CI value (circle: PG-C12; triangle: PG-C14; square: PG-C16; diamond: PG-C18:1). The values are expressed as mean ± SD (n = 3). Values with the same letter at each addition are not significantly different at p < 0.05.