Figures & data
Figure 1. A graphical representation of the framework for developing and evaluating environmental sampling activities. The 3 key questions, as well as the general answers are provided in the rings. Graphical depictions of technologies that may be employed are shown in the center, including (from top left):surface sampling, air sampling, water sampling, culture, protein detection, nucleic acid characterization, and real time aerosol monitoring (top center).
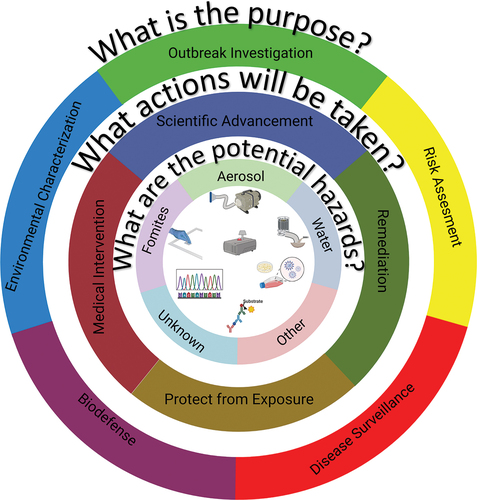
Table 1. Real-time bioaerosol measurement technologies, their operating principals, advantage and limitations.
Table 2. Overview of high-level analysis techniques for biological sampling identification. Sample recovery refers to the need to extract the sample from the collection substrate. *In some cases, bacteria and fungi can be collected onto growth media and cultivated from the collection surface. Analyte extraction refers to extracting the molecular analyte (typically nucleic acid or protein) from the recovered sample. **In some cases, proteins and nucleic acids can be detected from the collected sample without extraction or purification, particularly if surface proteins are the target or if extracellular DNA/RNA is present. Analyte conversion refers, in this case to the conversion of RNA to DNA for use in DNA amplification technologies. Analyte amplification refers to the amplification of the analyte to improve sensitivity. This applies most directly to sequencing application where a segment of DNA must be read multiple times (termed coverage) to ensure the sequence is accurate and the detection is confident. ***PCR and LAMP both implicitly amplify the analyte as a part of the detection process and can be used to amplify target DNA for downstream sequencing. Sensitivity refers to the fundamental sensitivity of the technique. Most nucleic acid detection techniques advertise single copy sensitivity, meaning that the technique can identify a single copy of the target sequence, but that does not account for instrument signal to noise, chemical inhibition or other factors that may decrease sensitivity.
