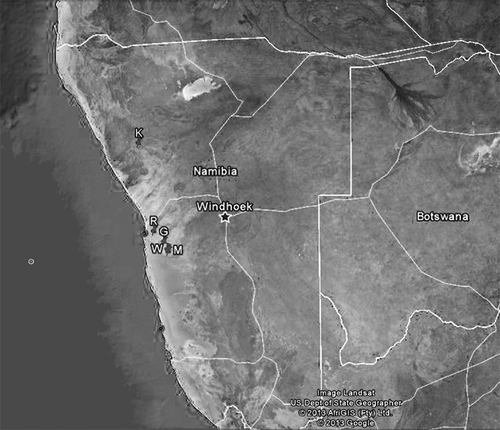
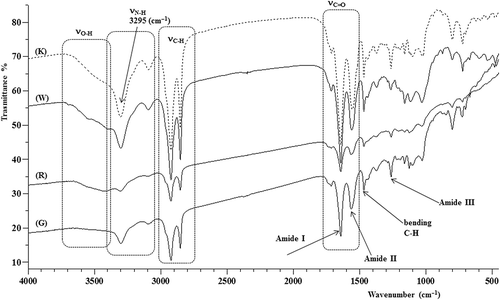
Figures & data
Table I. Granulometric soil analysis of the research stations. Values are expressed as percentages.
Table II. Arithmetic means and standard deviation of recorded parameters in different fieldwork stations.
Figure 2. Discriminant analysis. Plot of the first two canonical variables and centroids of the five populations.
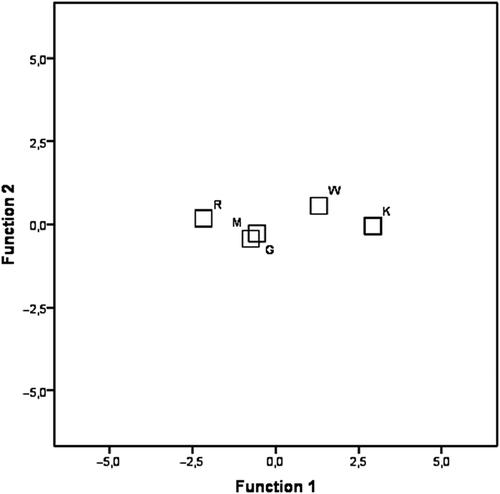
Figure 4. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrum of the M silk. Peaks spaced at 198 Da, 199 Da and 215 Da are labeled with the symbol *, whereas those spaced at 231 Da and 245 Da are marked with the symbol •.
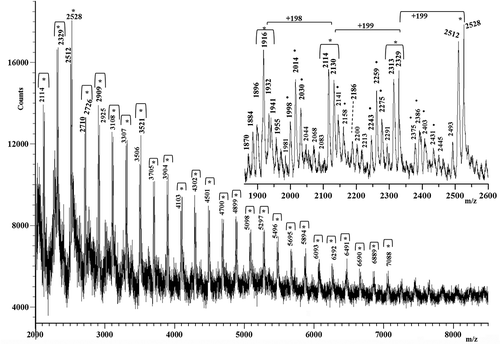
Figure 5. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrum of the K silk.
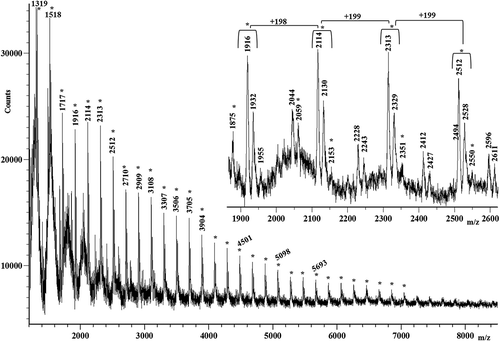
Figure 6. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrum of the R silk.
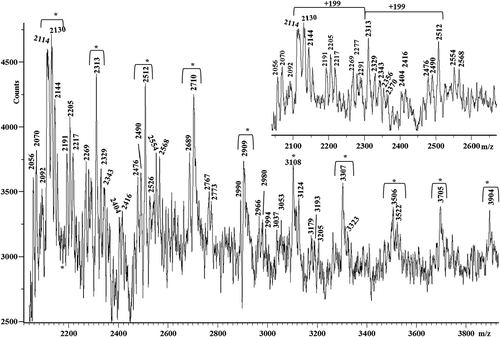
Figure 7. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectra in the higher mass range of M, K, R, W and G silks.
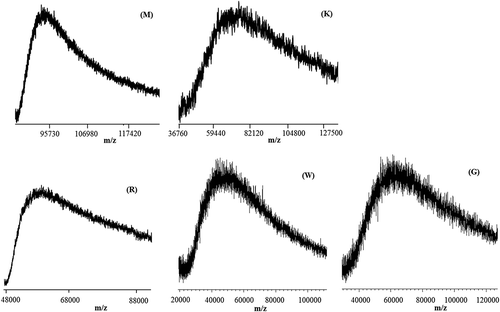
Figure 8. Differential scanning calorimetry (DSC) curves in the range from –70 to 180°C (recorded at 10 C/min) in the first heating run of the different silks.
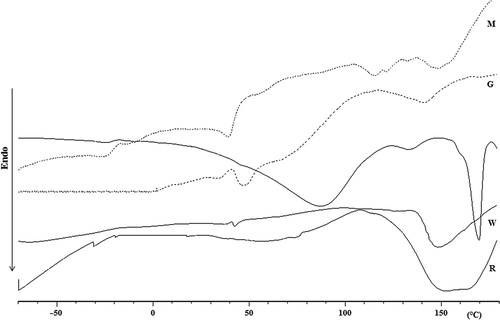
Table III. Mass differences of the most common amino acid sequences.
Table IV. Assignments of the most important peaks observed in the low mass range of the matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectra of the silks M, R and K. In parentheses are reported the variable amino acids sequences of each polypeptide.