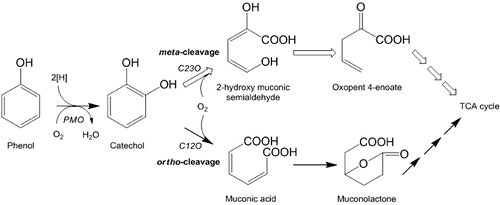
Figures & data
Table 1. Morphological, cultural and biochemical characteristics of isolated strains able to degrade phenol.
Figure 1. Growth kinetics of (A) IES-Ps-1, (B) IES-S and (C) IES-B in nutrient broth under shake flask conditions supplemented with phenol. Experimental data points are shown with dots. Haldane–Andrew regression curves are mentioned with regular lines and Luong–Levenspiel regression curves are indicated with dashed line.
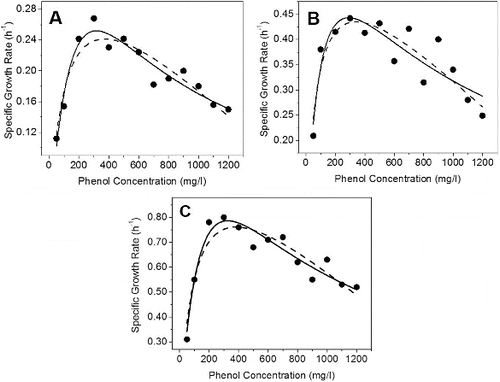
Table 2. Kinetic parameters obtained using Monod, Luong–Levenspiel and Haldane-Andrew models in batch mode with pure culture of Pseudomonas (IES-Ps-1), Pseudomonas (IES-S) and Basillus (IES-B) growing in NB supplemented with phenol.
Figure 2. Growth kinetics of phenol degradation by Pseudomonas IES-Ps-1 in MM medium supplemented with phenol. (A) 700 mg/L, (B) 900 mg/L and (C) 1050 mg/L.
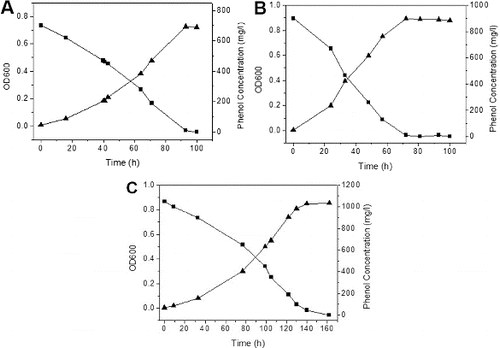
Figure 3. Cellular morphology of (A) Pseudomonas sp. IES-Ps-1 at 6500X magnification, (B) Bacillus sp. IES-B at 5500X magnification and (C) Pseudomonas sp. IES-S at 6500X magnification when grown in nutrient broth and the respective cells of IES-Ps-1, (D) IES-B (E) and IES-S (F) grown in mineral medium supplemented with phenol.
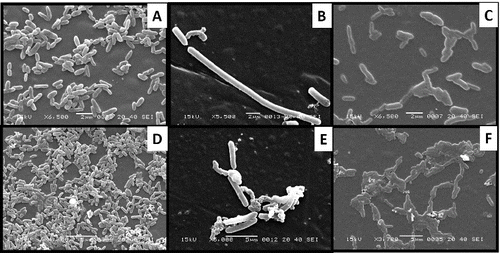
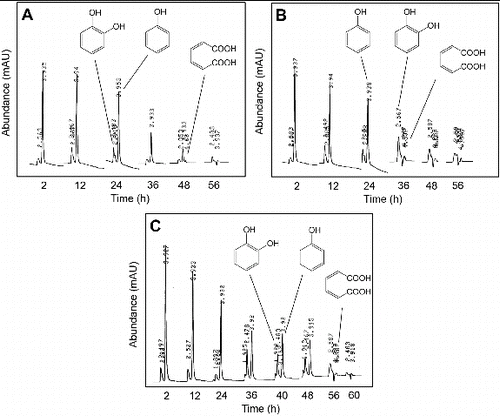
Figure 4. HPLC chromatograms showing the degradation of phenol in a bioreactor under non-sterile conditions supplemented with 700 mg/L of phenol when the bioreactor was inoculated with the cells of (A) Pseudomonas sp. strain IES-Ps-1, (B) Bacillus sp. strain IES-B and (C) Pseudomonas sp. strain IES-S.