Figures & data
Figure 1. A schematic diagram of the IRR-PCR strategy. Step 1, DNA fragments preparation: the segmented seven exons were amplified using gDNA templates and primers containing 5′-end overlapping homologous sequence with the adjacent exon by normal PCR. Step 2, isothermal assembly of fragments: after ExoI treatment and heat inactivation, the first-round PCR products were mixed and subjected to isothermal assembly. Step 3, PCR amplifying splicing products: the assembled full-length CDS was amplified by PCR with the outermost primers (F1 and R1). The PCR products of splicing DNA molecules could be directly sequenced or cloned into a linear functional plasmid by isothermal recombination or Ω-PCR method. The black blocks indicate the overlapping homologous sequences with the multiple cloning sites of prokaryotic expression plasmid pGEX-4T-2. The dashed box in step 2 indicates the detailed process of isothermal in vitro recombination reaction.
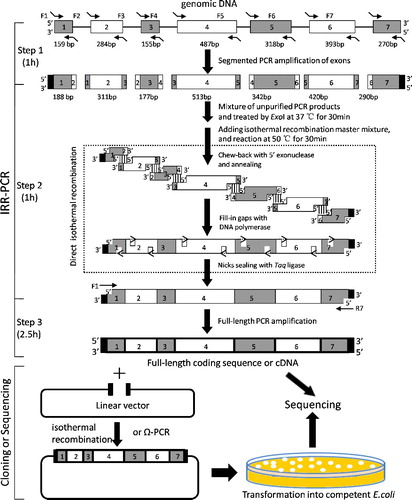
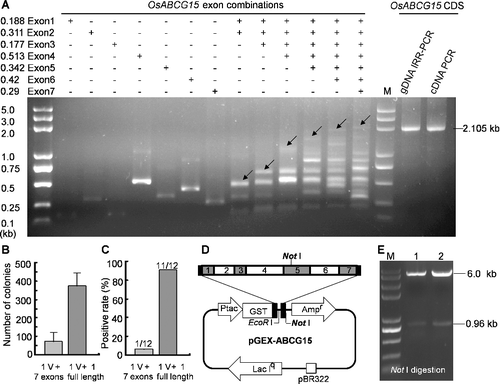
Figure 2. Splicing full-length CDS of OsABCG15 from gDNA by the IRR-PCR strategy. (a) PCR-amplified exon products and assembled products of different exons by isothermal in vitro recombination were displayed on agarose gel electrophoresis. The expected band joined by seven exons and other intermediate combination products were demonstrated. Arrows indicate the predicted target products of the exon combinations. The full-length CDS of OsABCG15 assembled by IRR-PCR had the same size with the anther-specific cDNA derived from conventional cDNA cloning. The amplified full-length sequence (2105 bp) contained a 2067-bp coding sequence and two short flanking sequences. (b) and (c) Comparison of transformation efficiency and positive rate between direct assembly and IRR-PCR strategy. Seven exons and one IRR-PCR splicing full length were assembled separately into the linear pGEX-4T-2 vector. (d) The map of recombinant plasmid pGEX-ABCG15 and (e) identification of positive clone by Not I digestion.