Figures & data
Figure 2. Plant responses employed to tackle with many important environmental stresses such as drought, cold, heat, salinity, pH and submergence. Drought and heat stresses often happen simultaneously leading to additive damages.
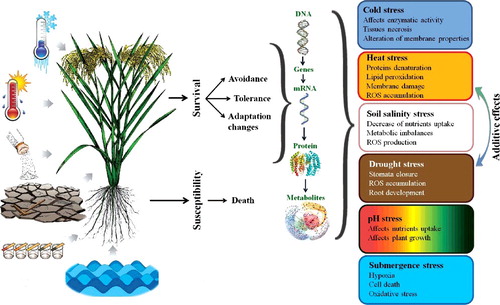
Figure 3. The function of BADH in the tolerance to abiotic stresses via the MAPK signalling pathway, and also metabolic synthesis of glycine betaine in plants (Modified from Yu et al. [Citation47]). Note: AAO, ABA-aldehyde oxidase; ABI, encoding gene for PP2C; APX, ascorbate peroxidase; CAT, catalase; CDPK, Ca2+-dependent calmodulin-independent protein kinase; CMO, choline monooxygenase; MAD, malondialdehyde; MAPK, mitogen-activated protein kinase; NAC, NAC transcription factor; NCED, 9-cis epoxycarotenoid dioxygenase gene; PA, phosphatidic acid; PLD, phospholipase D; POD, peroxidase; PP2C, protein phosphatase 2 C; REL, relative electrolyte leakage; RWC, relative water content; SnRK2, sucrose non-fermenting 1-related protein kinase 2; SOD, superoxide dismutase; WRKY, WRKY transcription factor.
![Figure 3. The function of BADH in the tolerance to abiotic stresses via the MAPK signalling pathway, and also metabolic synthesis of glycine betaine in plants (Modified from Yu et al. [Citation47]). Note: AAO, ABA-aldehyde oxidase; ABI, encoding gene for PP2C; APX, ascorbate peroxidase; CAT, catalase; CDPK, Ca2+-dependent calmodulin-independent protein kinase; CMO, choline monooxygenase; MAD, malondialdehyde; MAPK, mitogen-activated protein kinase; NAC, NAC transcription factor; NCED, 9-cis epoxycarotenoid dioxygenase gene; PA, phosphatidic acid; PLD, phospholipase D; POD, peroxidase; PP2C, protein phosphatase 2 C; REL, relative electrolyte leakage; RWC, relative water content; SnRK2, sucrose non-fermenting 1-related protein kinase 2; SOD, superoxide dismutase; WRKY, WRKY transcription factor.](/cms/asset/96b89cb0-c798-4ad7-a336-15cf80bbda4d/tbeq_a_1478748_f0003_c.jpg)