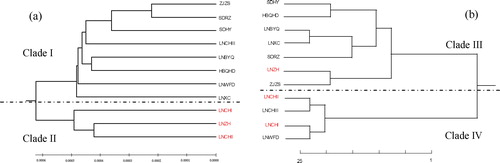
Figures & data
Figure 1. Haplotype distribution and relationships of the 11 populations of Glehnia littoralis. https://map.baidu.com/13438994,4328487,12z
Note: Circles represent sampling locations. H1, H2, and H3 represent different haplotypes detected.
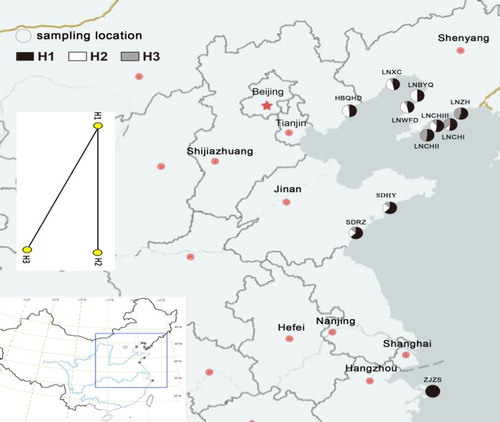
Table 1. Details of all the populations of Glehnia littoralis.
Table 2. RAPD primer sequences, number of loci and number of polymorphic loci.
Table 3. Information of variation loci of ITS haplotypes in Glehnia littoralis.
Table 4. Haplotype distribution and haplotype diversity in 11 populations of Glehnia littoralis.
Table 5. Genetic diversity of each population of Glehnia littoralis.