Figures & data
Table 1. Analysis of variance (ANOVA) of soybean seed yield per plant (YP) under drought stressed and full irrigation conditions for 149 soybean accessions.
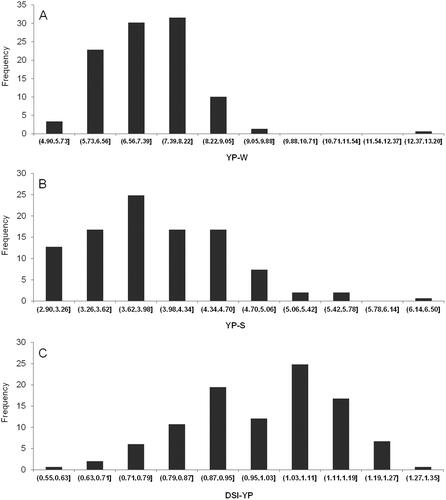
Table 2. Descriptive statistics of seed yield per plant (YP) under drought stressed and full irrigation conditions and DSI-YP values for 149 soybean cultivars.
Table 3. Phenotypic BLUP (best linear unbiased prediction) correlations between seed yield per plant (YP) under drought stressed and full irrigation conditions and DSI-YP in the 149 soybean cultivars.
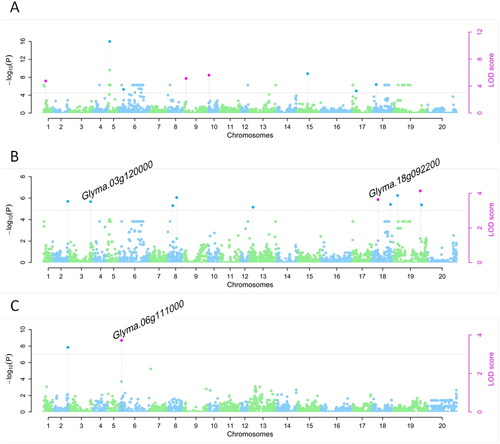
Table 4. SNPs significantly associated with YP under drought stressed and full irrigation conditions and DSI-YP.