Figures & data
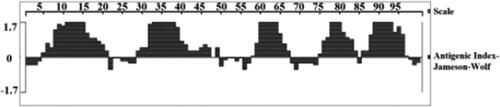
Figure 2. Basic chemical and physical features of SeCystatin. (A) Hydrophilicity and hydrophobicity regions predicted by Protscale. (B) Signal peptide analyzed by Singal P. (C) Transmembrane domain predicted by TMHMM.
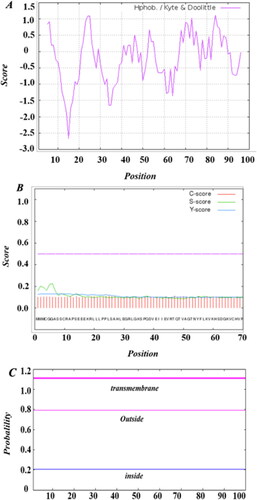
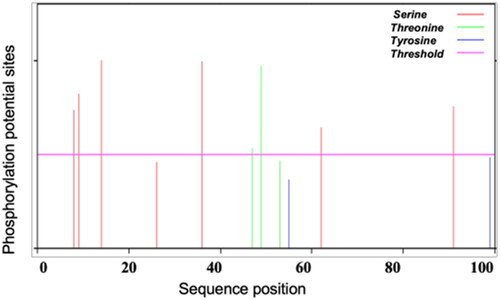
Table 1. Potential B-cell epitopes of SeCystatin predicted by DNAMAN.
Figure 5. Prediction of protein structure and model quality of SeCystatin. (A) Secondary structure composition predicted by PSIPRED. (B) Three-dimensional structure constructed by Phyre 2 based on homology modeling; the red arrow points to the QxVxG conserved domain. (C) Quality validation of homology modeled SeCystatin structures by Errat. Yellow bar indicates protein regions with misfolding at 95% confidence level.
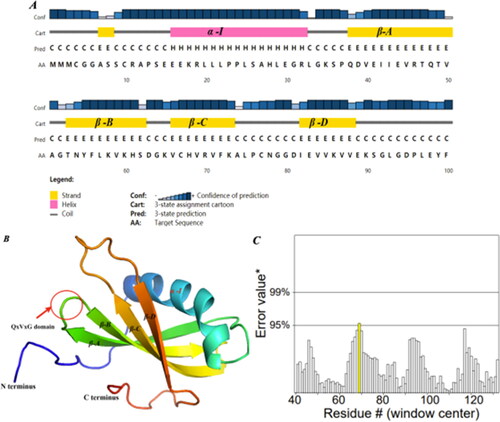
Figure 6. Multiple sequence alignments of cystatin. Different colored columns represent conserved sites among cystatin sequences. The letters in the consensus sequence represent the identical amino acid residues in the compared sequences. "ID%" refers to the percentage of similarity between each sequence and SeCystatin. All sequences were aligned using Clustal Omega and edited using Jalview.
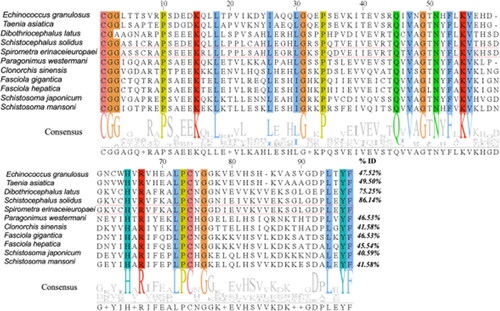
Figure 7. Phylogenetic evaluation of cystatins from different parasite species. The evolution tree was constructed using MEGA-X and further edited by Evolview; the green bar on the right side of the figure represents the location and size of the conserved cystatin domain.
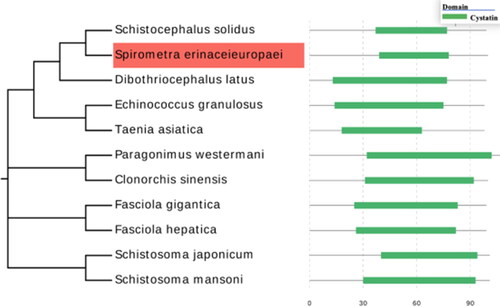
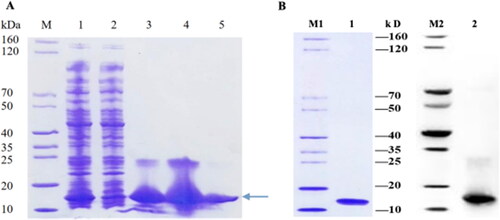
Figure 8. Expression, purification, and verification of SeCystatin. (A) Expression and purification results of SeCystatin analyzed by SDS-PAGE. Lane M: SDS-PAGE protein marker (cat. no. MY1397, MerryBio, Nanjing, China). Lane1: Supernatant after centrifugation of whole bacteria. Lane 2: Supernatant after incubation with Ni-IDA for 90 min. Lanes 3–4: Elution fraction of 50 mmol/L imidazole. Lane 5: Elution fraction of 300 mmol/L imidazole. (B) Quality and specificity of SeCystatin were verified by SDS-PAGE and Western blot analysis. Lane 1-2: Purified SeCystatin. M1: SDS-PAGE protein marker (cat. no. MY1397, MerryBio, Nanjing, China). M2: Western blot marker (cat. no. MY00521, MerryBio, Nanjing, China).
Data availability statement
The datasets analyzed in this study are available from the corresponding author upon reasonable request.