Figures & data
Figure 1. Overview of the three retinal isoforms of CRB1 and its location. All isoforms share exon 6. CRB1-B is the most abundant retinal isoform. It has a unique N-and C-terminus while sharing the transmembrane domain as well as a large part of the extracellular domain with CRB1-A. the patient’s novel nonsense mutation affects CRB1-A and CRB1-B, while the missense mutation affects all retinal CRB1 isoforms. CRB1 is thought to play an important role in maintaining the integrity of the OLM.
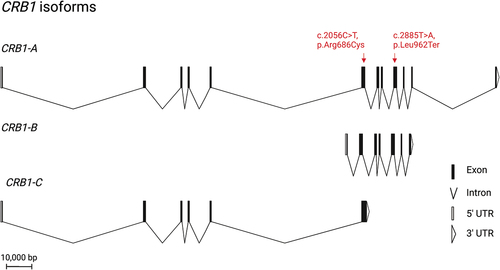
Figure 2. Clinical phenotype of patient with compound heterozygous for single nucleotide variants in CRB1. (a) Infrared (left) and autofluorescence (right) imaging shows atrophic changes in the central retina with hyperfluorescent spots outside the temporal arcades. (b) Characteristic for a CRB1-associated RP, both left and right eye show increased retinal thickness. the retinal lamination pattern is coarse, reminiscent of an immature retina and disrupted by intraretinal exudates.
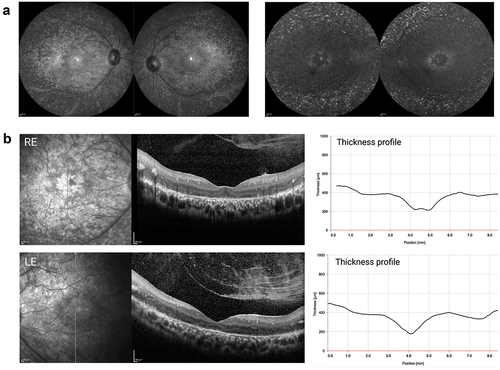
Table 1. Classification of the novel CRB1 c.2885T>A (p.Leu962Ter) and CRB1 c.2056C>T (p.Arg686Cys) variant using ACMG standards and guidelines.
Figure 3. Therapeutic “read-through” approach mediated by CRISPR-pass. While base editing with CRISPR-pass does not allow a true ribosomal read-through, nonsense mutations not amenable to conventional base editing strategies can be turned into a missense change by using an ABE-xCas base editing construct (CRISPR-pass), targeting the coding or non-coding strand. in this case, a bystander mutation would occur when targeting the coding strand due to the presence of an additional a in the editing window. the tryptophan and glutamine missense changes resulting from CRISPR-pass mediated editing are predicted to be “possibly damaging” by the in-silico tool Polyphen 2, which evaluates the functional impact of missense changes.
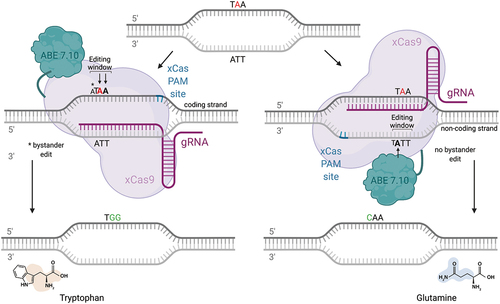
Figure 4. Overview of base editing strategy for a C>T transition mutation. the patient could be transferred to a carrier status by using an ABE7.10 or an ABE8 fused to a SaCas9-KKH, which displays a relaxed PAM site 5’-NNNRRT-3’ and an editing window within 2–15 nucleotides upstream of the PAM site. No additional A’s are present in the editing window, greatly reducing the possibility of bystander mutations.
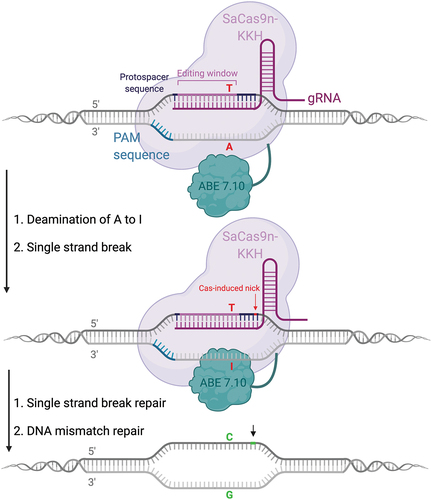
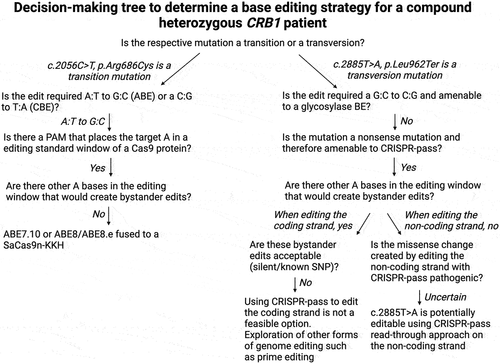
Figure 5. Decision making tree for developing a base editing strategy for a patient with compound heterozygous mutation in CRB1.