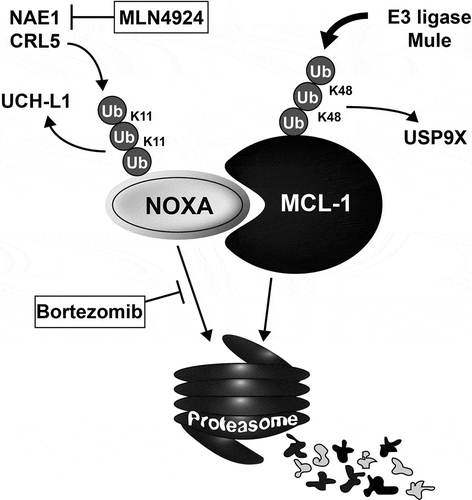
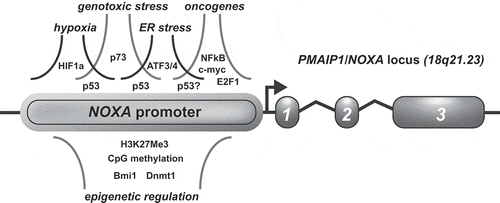
Figures & data
Table 1. Ordering of cell stress pathways that affect Noxa level or function, with potential drugs and stage of clinical development indicated.
![Figure 1. Functional interactions among Bcl-2 family members.Inhibitory interactions among pro-survival members (dark grey boxes), and strong (dark grey ovals) versus weak (light grey ovals) BH3-only members. Stimulatory (arrow) and inhibitory (blockline) interactions among pro-survival Bcl-2 members, BAX/BAK1, and strong BH3-only members. NOXA is tentatively represented as activator of BAK and BAX based on Refs. 20–23.Figure compiled and adapted from information and similar schemes represented in Refs [Citation8,Citation9,Citation18,Citation20].](/cms/asset/83f5cab4-f964-4fd9-bbe8-ca7ea3246fa5/iett_a_1349754_f0001_b.gif)