Figures & data
Figure 1. Insights into the molecular recognition of the minimal GalNAc moiety by the CLEC10A CRD domain by NMR and X-Ray crystallography. (a) 1H,15N-HSQC spectra of CLEC10A in the apo state (blue) and with the addition of 0.5 (black) and 5 equivalents (red) of α-MeGalNAc at 600 MHz and 310 K. The residues with asterisks (*) correspond to those that are not present in the apo state and appear in the bound state. In this technique, the resonances of the most affected residues by the presence of α-MeGalNAc are perturbed. (b) Atomic fluctuation (Cα) analysis of CLEC10A CRD domain in the apo state (left) and bound to GalNAc (right) derived from 100 ns molecular dynamics (MD) simulations [Citation20]. The data correspond to the average protein structure derived from the MD simulations. (c) Chemical shift perturbation (CSP) analysis to quantify the perturbation in the resonances observed in A. Two cut-off lines at 0.04 ppm and 0.08 ppm shown in black and blue dash lines, respectively, and are used to differentiate strong and moderate shift in the presence of α-MeGalNAc. (d) X-ray crystallography structure (PDB code: 6PY1) [Citation21] of CLEC10A CRD domain complexed with α-MeGalNAc with the most affected residues determined in panel C highlighted in red. An expansion of the binding-site of CLEC10A (gray carbons) and α-GalNAc (yellow carbons) in stick representation containing the Q267/P268/D269 and W291/N292/D293 motifs and the residues Y236, H286, H284, W271 and E280. The calcium ion (Ca2+) that coordinates OH3 and OH4 of α-GalNAc is shown with a purple sphere. The 3D models in panels b and d were created using Pymol 2.4.1 [Citation22].
![Figure 1. Insights into the molecular recognition of the minimal GalNAc moiety by the CLEC10A CRD domain by NMR and X-Ray crystallography. (a) 1H,15N-HSQC spectra of CLEC10A in the apo state (blue) and with the addition of 0.5 (black) and 5 equivalents (red) of α-MeGalNAc at 600 MHz and 310 K. The residues with asterisks (*) correspond to those that are not present in the apo state and appear in the bound state. In this technique, the resonances of the most affected residues by the presence of α-MeGalNAc are perturbed. (b) Atomic fluctuation (Cα) analysis of CLEC10A CRD domain in the apo state (left) and bound to GalNAc (right) derived from 100 ns molecular dynamics (MD) simulations [Citation20]. The data correspond to the average protein structure derived from the MD simulations. (c) Chemical shift perturbation (CSP) analysis to quantify the perturbation in the resonances observed in A. Two cut-off lines at 0.04 ppm and 0.08 ppm shown in black and blue dash lines, respectively, and are used to differentiate strong and moderate shift in the presence of α-MeGalNAc. (d) X-ray crystallography structure (PDB code: 6PY1) [Citation21] of CLEC10A CRD domain complexed with α-MeGalNAc with the most affected residues determined in panel C highlighted in red. An expansion of the binding-site of CLEC10A (gray carbons) and α-GalNAc (yellow carbons) in stick representation containing the Q267/P268/D269 and W291/N292/D293 motifs and the residues Y236, H286, H284, W271 and E280. The calcium ion (Ca2+) that coordinates OH3 and OH4 of α-GalNAc is shown with a purple sphere. The 3D models in panels b and d were created using Pymol 2.4.1 [Citation22].](/cms/asset/1c664747-b9f6-43b2-a997-c6247be87935/iett_a_2374743_f0001_oc.jpg)
Figure 2. CLEC10A stimulation of human monocyte-derived dendritic cells. (a) Without toll-like receptor (TLR) activation, CLEC10A stimulation using anti-CLEC10A antibodies, Tn-glycosylated peptides or Tn-dendrimers induces a signaling cascade comprising of ERK1/2, p90RSK, and CREB, with possible contributions of Syk, PLCγ2, PKCδ, p38, MEK1/2, JNK, Akt, and β-catenin [Citation30,Citation32]. This signaling cascade activates a distinct transcriptional program [Citation33]. Furthermore, the CLEC10A stimulation reduces the glycolysis in monocyte-derived dendritic cells [Citation33]. The stimulated dendritic cells produce increased amounts of IL-10, IL-6 and TNF-α and reduce the proliferation of target CD4+ T cells [Citation30]. (b) In combination with TLR stimulation, the CLEC10A-stimulated signaling cascade includes p38, MEK1/2, ERK1/2, p90RSK, and CREB [Citation18]. The transcription of the cytokine genes is initiated and this leads to increased production and secretion of IL-10, IL-6 and IL-8 [Citation18,Citation19]. Besides reducing their proliferation, the IL-10 drives the differentiation of CD4+ T cells into type 1 regulatory T (Tr1) cells. These cells produce IL-10 themselves and can repress the activation of responder CD4+ T cells [Citation31]. (c) CD45 on activated CD4+ and CD8+ T cells can directly bind CLEC10A, which induces apoptosis in the T cells [Citation12,Citation34].
![Figure 2. CLEC10A stimulation of human monocyte-derived dendritic cells. (a) Without toll-like receptor (TLR) activation, CLEC10A stimulation using anti-CLEC10A antibodies, Tn-glycosylated peptides or Tn-dendrimers induces a signaling cascade comprising of ERK1/2, p90RSK, and CREB, with possible contributions of Syk, PLCγ2, PKCδ, p38, MEK1/2, JNK, Akt, and β-catenin [Citation30,Citation32]. This signaling cascade activates a distinct transcriptional program [Citation33]. Furthermore, the CLEC10A stimulation reduces the glycolysis in monocyte-derived dendritic cells [Citation33]. The stimulated dendritic cells produce increased amounts of IL-10, IL-6 and TNF-α and reduce the proliferation of target CD4+ T cells [Citation30]. (b) In combination with TLR stimulation, the CLEC10A-stimulated signaling cascade includes p38, MEK1/2, ERK1/2, p90RSK, and CREB [Citation18]. The transcription of the cytokine genes is initiated and this leads to increased production and secretion of IL-10, IL-6 and IL-8 [Citation18,Citation19]. Besides reducing their proliferation, the IL-10 drives the differentiation of CD4+ T cells into type 1 regulatory T (Tr1) cells. These cells produce IL-10 themselves and can repress the activation of responder CD4+ T cells [Citation31]. (c) CD45 on activated CD4+ and CD8+ T cells can directly bind CLEC10A, which induces apoptosis in the T cells [Citation12,Citation34].](/cms/asset/610e1129-1dbe-4a2d-86b6-153c1dd34dd7/iett_a_2374743_f0002_oc.jpg)
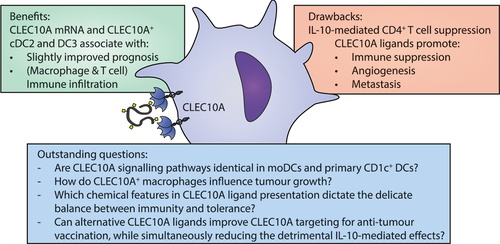
Figure 3. Benefits, drawbacks and outstanding questions for CLEC10A targeting in the tumor microenvironment.