Figures & data
Figure 1 Alcohol dehydrogenases (ADH) and short-chain dehydrogenase/reductase catalyse the oxidation of retinol to retinaldehyde, which is subsequently oxidised by aldehyde dehydrogenases (ALDH) to retinoic acid. Retinoic acid, all-trans (atRA) and 9-cis (9cRA) isomers are further metabolised by cytochrome P450 enzymes (including CYP26A1, B1 and C1) to inactive polar metabolites resulting in retinoid excretion.
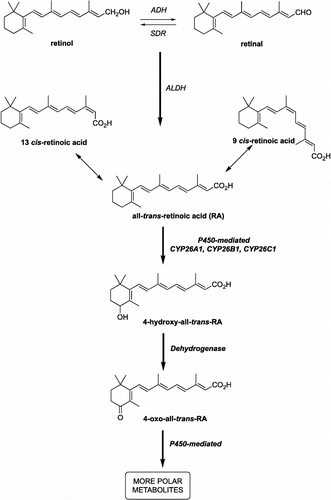
Table I. P450 sequences identified by PSI-BLAST search using CYP26A1 as the query sequence.
Figure 2 CYP26A1 sequence and ClustalW (1.82) alignment with CYP2C9, CYP2C8 and CYP3A4. The colour coding of amino acid type: red, small + hydrophobic (AVFPMILWY); blue, acid (DE); magenta, basic (RHK); green, hydroxyl + amine + basic (STYHCNGQ). Substrate recognition sites (SRS) and conserved P450 domains (ETLR, PERF and Haem binding domain) are indicated. “*” means that the residues are identical, “:” means that conserved substitutions have been observed, “.” means that semi-conserved substitutions are observed.

Table II. Validation results for the lowest energy CYP26A1 models (produced from the three different templates) and the 3D structural templates.
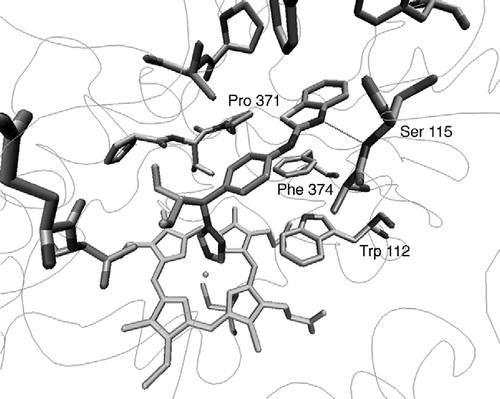
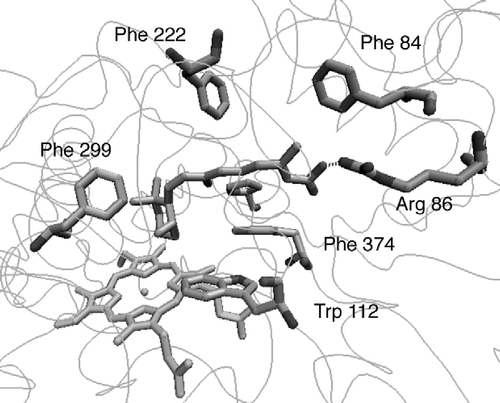
Figure 3 CYP26A1 model active site (a) before and (b) after active site optimisation with the (S)-R115866 bound inhibitor.
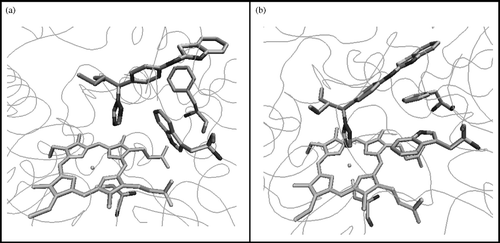
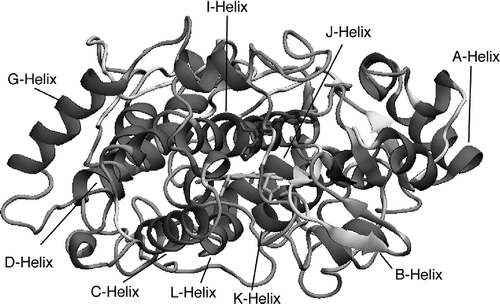
Table III. 3. Comparison of CYP26A1 model and CYP3A4 template secondary structure.