Figures & data
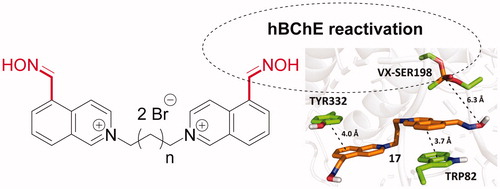
Scheme 2. Preparation of quaternary salts 16–26. Reagents and conditions: (b) CH3I, EtOH, reflux, 24 h, 71%; (c) dibromoalkanes, DMF, 73 °C, 48 h, 33–94%.
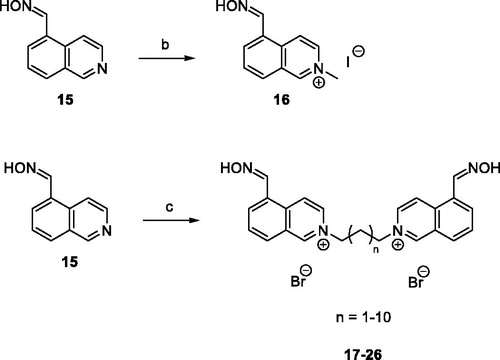
Scheme 3. Preparation of bisquarternary salts 30–33. Reagents and conditions: (d) DMF, 73 °C, 48 h, 53–77%; (e) bis(chloromethyl)ether, DMF, 73 °C, 48 h, 45%
Table 1. Inhibition of hAChE and hBChE by prepared compounds.
Table 2. Reactivation of hAChE inhibited by sarin, VX and paraoxon by selected compounds.
Table 3. Reactivation of hBChE inhibited by sarin, VX and paraoxon by selected compounds.
Table 4. Reactivation kinetics of human recombinant BChE inhibited by NIMP (sarin surrogate) and NEMP (VX surrogate) using selected compounds.
Figure 3. Overlaid predicted binding modes of compounds 25 (yellow) and 26 (light pink) in hAChE (pdb id: 4ey7, left) and hBChE (pdb id: 4bds, right). The residues interacting with 25 and 26 are coloured in light green and purple, respectively.
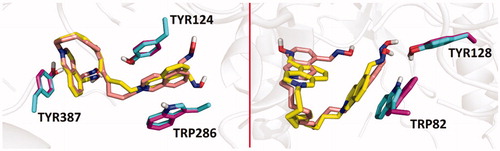
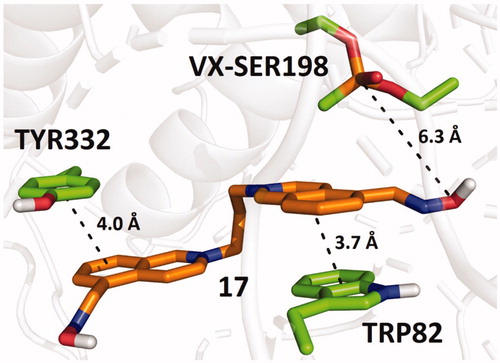
Figure 4. Predicted binding mode of compound 30 (blue) in hAChE inhibited by sarin (A; pdb id: 5fpq), VX (B; pdb id: 6cqt) and paraoxon (C; pdb id: 5hf9).
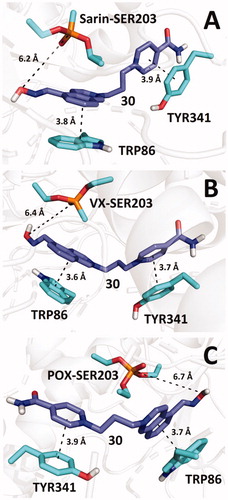
Table 5. The lowest predicted binding energies of the studied compounds in selected cholinesterase models by molecular docking.