Figures & data
Figure 1. Structure of selective oestrogen receptor blocker Tamoxifen (1), Steroidal aromatase inhibitors Exmestane (2), nonsteroidal aromatase inhibitor letrozole (4), anastrozole (5) Kinase inhibitors, Vemurafenib (6), Encorafenib (7), Non selective NSAIDs, Indomethacin (8), selective COX-2 inhibitors celecoxib (9), and 1,2,4 triazoles with anti-cancer activities (10) and anti-cancer 1,2,4 triazoles (10), (11), and B-RAFV600E inhibitor (12).
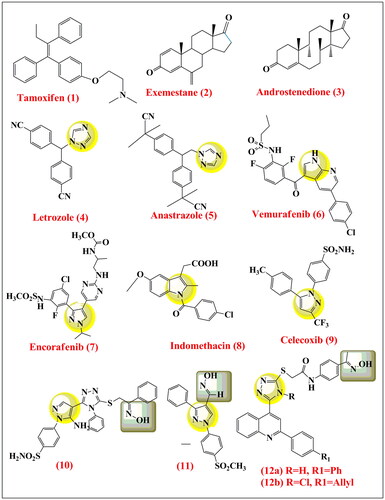
Figure 2. structural hyperdization of designed compounds from COX-2 inhibitors celecoxib (9) and anti-cancer 1,2,4 triazoles (10), (11), B-RAFV600E inhibitors (12a&b).
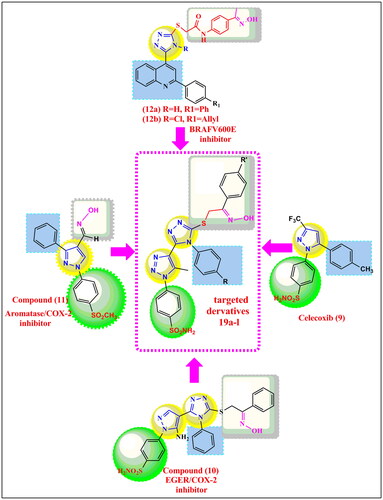
Table 1. Cell cycle analysis of compounds 19c, 19f, 19h, 19 l and control for breast MCF7 cells.
Table 2. Apoptosis of compounds 19c, 19f, 19h, 19 l and control against breast MCF7 cells.
Scheme 1. Reagents and conditions: (a) ethanol 95% reflux 4h., (b) alcoholic NaOH reflux 4h., (c) acetonitrile, triethylamine, reflux 10h., (d) acetonitrile, hydroxylamine hydrochloride, sod. acetate, reflux, 10h.
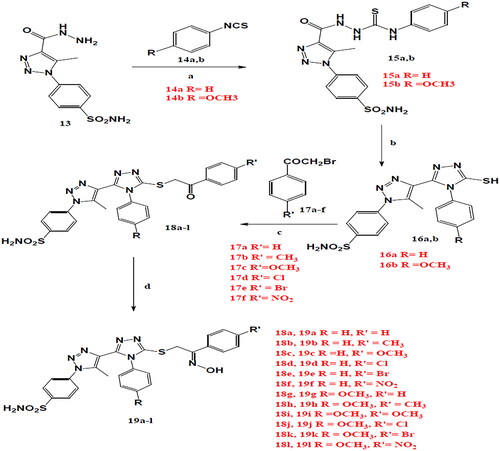
Table 3. In vitro COX-1 and COX-2 inhibitory activity of triazole derivatives 19a-l and reference drug celecoxib.
Figure 3. Cell viability % of compounds 19a-l and reference compound single dose (10 μM) against MCF-7, HEP-38, HCT-116 and A549 cancer cell line.
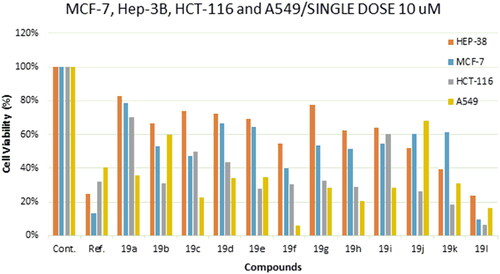
Figure 4. Compounds 19c, 19f, 19h, 19 l effect on DNA-ploidy flow cytometric analysis of breast MCF7 cells compared to negative control.
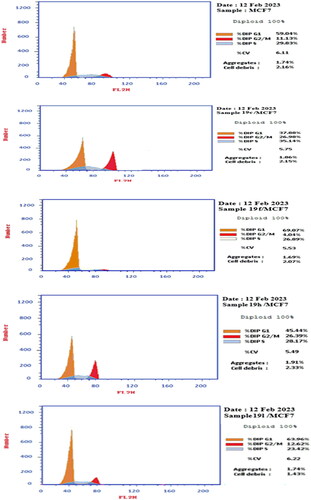
Figure 5. The percentage of Annexin-V-FITC-positive staining in breast MCF7 cells when treated with compounds 19c, 19f, 19h, 19 l compared to sorafenib and negative control.
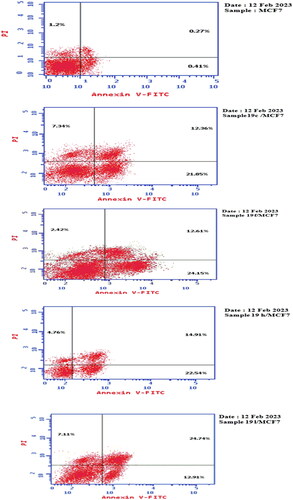
Figure 6. IC50% values of compounds 19c, 19f, 19h, 19 l compared to letrozole on aromatase against breast MCF7 cells.
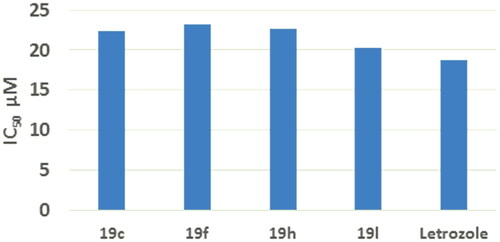
Table 4. In vitro MCF-7, HEP-3B, HCT-116, A549 and F180 inhibitory activity of triazole derivatives 19c,e,f,h,j-l and reference drugs doxorubicin, sorafenib, 5-F.U. and tamoxifen..
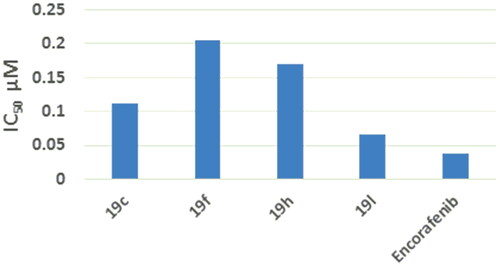
Figure 8. IC50 values of compounds 19c, 19f, 19h, 19 l compared to that of encorafenib on B-RAFV600E.
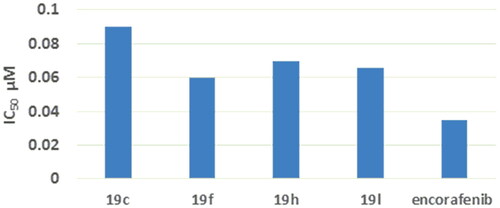
Table 5. % of NO release of the synthesised compounds.
Figure 9. (A) Visual representation (3D) of celecoxib docked with 3LN1 active site (B) Visual representation (3D) of compound 19c docked with 3LN1active site, (C) Visual representation (3D) of compound 19f docked with 3LN1 active site (D) Visual representation (3D) of compound 19h docked with 3LN1 active site (E) Visual representation (3D) of compound 19 l docked with 3LN1 active site.
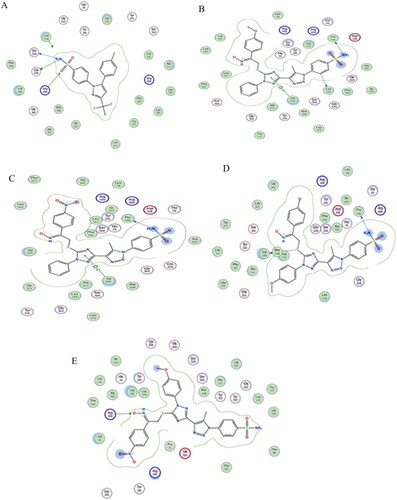
Table 6. Docking data of triazoles compounds 19a-l and celecoxib with COX-2 active site (3LN1).
Figure 10. (ِA) Visual representation (2D) of compound 19c docked with 1M17 active site, (B) Visual representation (2D) of compound 19f docked with 1M17 active site (C) Visual representation (2D) of compound 19h docked with 1M17 active site (D) Visual representation (2D) of compound 19 l docked with 1M17 active site (E)Visual representation (2D) of encorafenib docked with 1M17 active site (F) Visual representation (2D) of 4-anilinoquinazline docked with 1M17 active site.
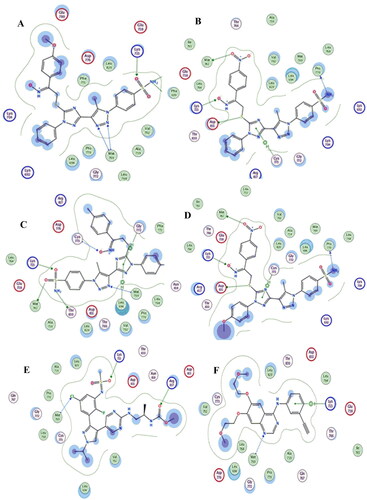
Table 7. Docking data of triazoles 19a-l and encorafenib with EGFR enzyme (1M17).
Figure 11. (A) Visual representation (3D & 2D) of letrozole docked with aromatase enzyme 3EQM active site (B) Visual representation (3D & 2D) of compound 19c docked with aromatase enzyme 3EQM active site, (C) Visual representation (3D & 2D) of compound 19f docked with aromatase enzyme 3EQM active site (D) Visual representation (3D & 2D) of compound 19h docked with aromatase enzyme 3EQM active site (E) Visual representation (3D & 2D) of compound 19 l docked with aromatase enzyme 3EQM active site.
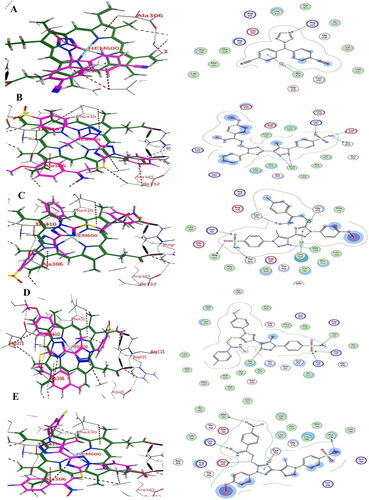
Table 8. Docking data of triazoles 19a-l and letrozole with aromatase enzyme (3EQM).
Figure 12. (A) Visual representation (2D) of compound 19c docked with enzyme B-RAFV600E (PDB entry 1UWJ) active site, (B) Visual representation (2D) of compound 19f docked with B-RAFV600E (C) Visual representation (2D) of compound 19h docked with B-RAFV600E (D) Visual representation (2D) of compound 19 l docked with B-RAFV600E (E)Visual representation (2D) of encorafenib docked with B-RAFV600E (F) Visual representation (2D) of Sorafenib docked with B-RAFV600E.
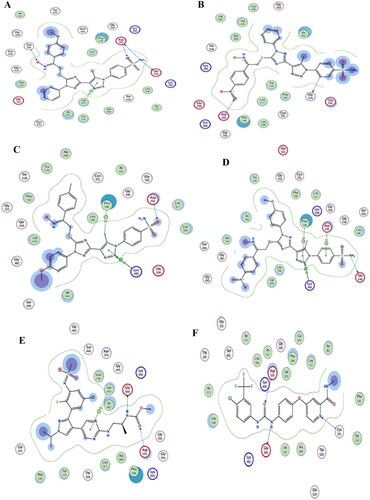
Table 9. Docking data of the most active triazoles 19c,f,h,l and encorafenib kinase enzyme B-RAFV600E (PDB, ID: 1UWJ) active site.