Figures & data
Table 1. Comparison of H1 and H2.
Figure 1. A family of grain scans involving a parameter β to illustrate differences between H1 and H2. Top, from left to right: Grain scans for ,
, and
. Grain
(orange [light gray]) fills the square
,
(blue [gray]) fills the rest of the unit square. The bottom row shows the computed ellipses
with
for i = 1, 2.
![Figure 1. A family of grain scans involving a parameter β to illustrate differences between H1 and H2. Top, from left to right: Grain scans for β=0.1, β=0.5, and β=0.9. Grain G1 (orange [light gray]) fills the square [0,β]×[0,β], G2 (blue [gray]) fills the rest of the unit square. The bottom row shows the computed ellipses ci+BAi2 with Ai=(Σ(Gi))−1 for i = 1, 2.](/cms/asset/e22424c1-c71c-4d84-8438-032b0a35874b/tphm_a_2180679_f0001_oc.jpg)
Figure 2. Ratios of the areas of the grain and the corresponding ellipsoid for i = 1 (orange [light gray]) and i = 2 (blue [gray]) as a function of β.
![Figure 2. Ratios νi/vol(BAi2) of the areas of the grain and the corresponding ellipsoid for i = 1 (orange [light gray]) and i = 2 (blue [gray]) as a function of β.](/cms/asset/878b484e-131b-434f-b924-d82816560e71/tphm_a_2180679_f0002_oc.jpg)
Figure 3. Diagrams for the family of grain scans from obtained via H1 (top row) and H2 (bottom row).

Figure 4. Ratio of the area of the original grain and the area of the corresponding diagram cell obtained by H1 (left) and H2 (right) for i = 1 (orange [light gray]) and i = 2 (blue [gray]) as a function of β.
![Figure 4. Ratio of the area νi of the original grain and the area of the corresponding diagram cell obtained by H1 (left) and H2 (right) for i = 1 (orange [light gray]) and i = 2 (blue [gray]) as a function of β.](/cms/asset/47422b7e-9631-460c-8ee4-92c50e725ef4/tphm_a_2180679_f0004_oc.jpg)
Figure 5. Accuracy (within the cross-validation paradigm) of the interpolation/extrapolation model plotted for each using polynomials of degree 1, 2, and 3, respectively for the fit.
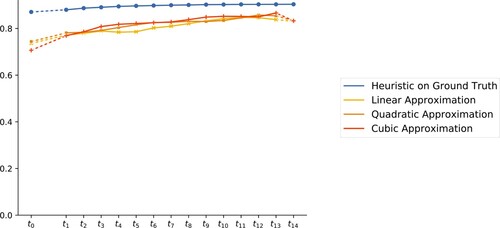
Figure 6. Visual comparison of diagrams obtained from full data (available for ) and the interpolation/extrapolation model that uses all data except for the respective moment in time. A fixed 2D slice through the 3D data set at times
,
,
and
(top row); the corresponding diagram representation obtained via H1 (2nd row) and, respectively, via the interpolation/extrapolation model that uses only data from the other moments in time and best-fitting quadratic polynomials (3rd row); symmetric difference between the images from the 2nd and 3rd row (bottom row).
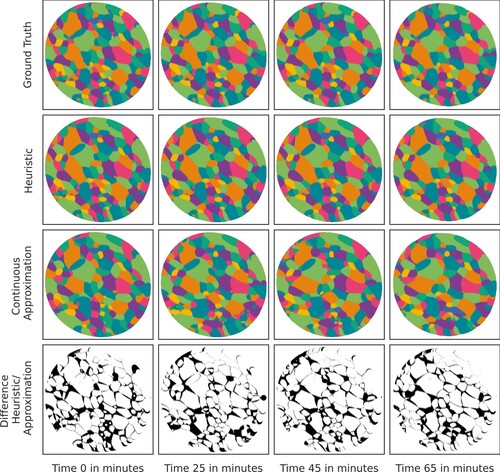
Table 2. Variance of for
of persistent grains.
Table 3. Variance of for
of non-persistent grains.
Table 4. Coefficients estimated by maximum likelihood estimation and their p-values for the described logistic regression model.
