Figures & data
Figure 1. (A) Western blot analysis of Bcl-2 family protein expression between an acquired model of ABT-737 Resistance (CHLA-15-ABTR) and the parent cell line, CHLA-15. (B) Co-IP showing Bim bound to Bcl-2 in CHLA-15 and to Mcl-1 in CHLA-15-ABTR. (C) Receptor tyrosine kinase human phosphoprotein microarray identifies differences in RTK expression in CHLA-15-ABTR vs. CHLA-15; RTK's blotted for are represented in duplicate side by side dots on the membrane for each RTK; ***; white stars designate positive controls. (D) Western blots of cells grown at steady-state confirm increased EGFR, pEGFR and downstream EGFR effectors in CHLA-15-ABTR.
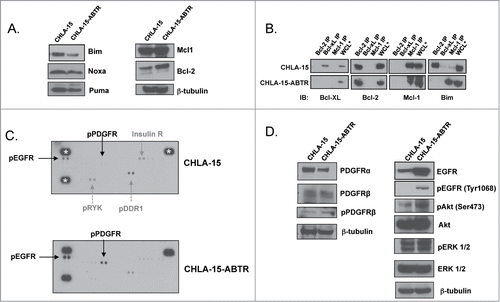
Figure 2. (A) Different subpopulations of SMS-SAN selected for ABT-737 resistance (SAN-ABTR) at different times from the Bcl-2 dependent SMS-SAN growing in > 150 nM of ABT-737 (parent cell line IC50 5 nM). (B) CHLA-15-ABTR cells were transfected with EGFR siRNA for 24 hours, then treated with ABT-737 for 24 additional hours at which time cell viability was assessed by WST-1. Mcl-1 dependent CHLA-15-ABTR (C) and Bcl-2 dependent CHLA-15 (D) were treated with erlotinib and ABT-737 simultaneously and assessed for changes in viability at 48 hours by WST-1 assay. *; statistically significant difference, p< 0.05, 95% Ci.
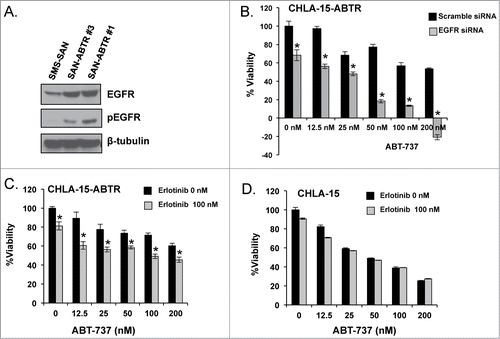
Figure 3. (A) WST-1 evaluation for survival following a 48-hour treatment with chemotherapy. Western blot analysis of EGFR signaling (B) and Bcl-2 family (C) proteins of untreated whole cell lysate from Bcl-2 dependent SMS-SAN and Mcl-1 dependent SK-N-BE(2). (D) Western analysis for EGFR and pEGFR performed in a panel of HR NB cell lines previously characterized for Bim binding patterns (Bcl-2 or Mcl-1 dependenceCitation4). Paired NB cell lines derived from the same patient's tumor at diagnosis and following cytotoxic therapy and relapse, with relapsed cell line designated with a *; SK-N-BE(1)/SK-N-BE(2)*, CHLA-15/CHLA-20*. NLF was incubated for 48 hours in the presence (+ FBS) or absence (- FBS) of serum and evaluated by western for EGFR and pEGFR.
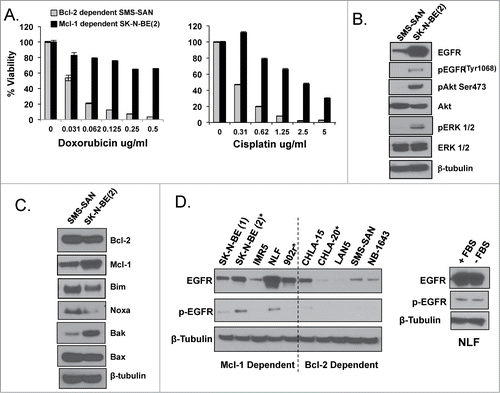
Figure 4. (A) Western analysis of EGFR protein expression with densitometry analyzed EGFR normalized to β-tubulin expression for SK-N-BE(2) wild type verses shEGFR-transfected cell lines. (B) Co-IP of pro-survival Bcl-2 proteins followed by immunoblot for pro-survival proteins and Bim, showing Bim moves from Mcl-1 over to the binding pocket of Bcl-2 following EGFR inhibition. (C) Western blot analysis for changes in Bcl-2 family and EGFR signaling proteins following EGFR knockdown in SK-N-BE(2)-shEGFR.
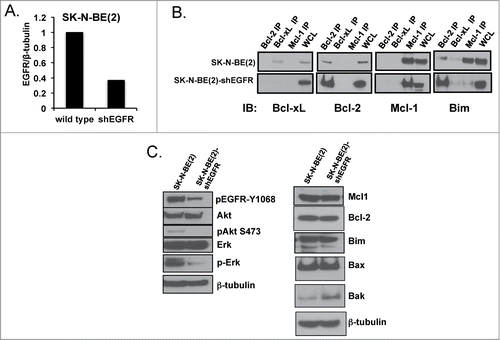
Figure 5. SK-N-BE(2) cells become exquisitely sensitive to both ABT-737 (A) and cytotoxic drugs (B & C) compared to scramble shRNA-transfected SK-N-BE(2) cells. Viability was assessed using WST-1 assay after 48 hours of incubation. All experiments were performed in technical triplicate and standard deviations represent the average of 2 separate biologic experiments. Standard deviations not shown have a less than 5% difference between biologic replicates.
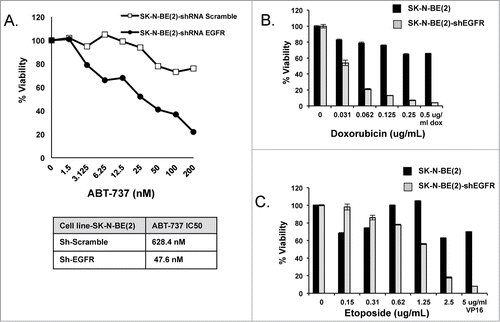
Figure 6. Mcl-1 dependent NB cells NLF (A) and SK-N-BE(2) (B) were exposed to EGFR inhibitors, erlotinib or cetuximab, and ABT-737 simultaneously and evaluated after 48 hours for changes in survival by WST-1. (C) NLF cells were exposed to given concentrations of U0126, erlotinib, or LY294002 for 24 hours, harvested for protein, and evaluated for changes in EGFR signaling and Bcl-2 family protein expression. (D) Protein from an Mcl-1 co-IP was treated with Lambda Protein Phosphatase (LPP) and then evaluated for changes in Bim:Mcl-1 interactions by immunoblot.
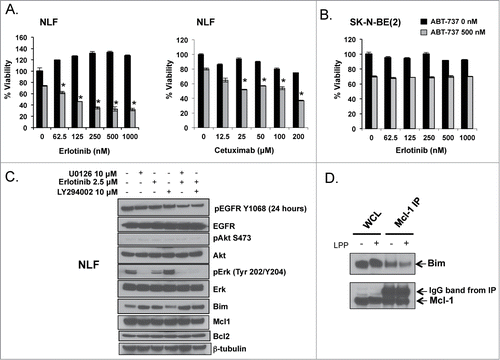
Figure 7. A. NLF was treated with UO126 (10 μM), LY294002 (2.5 μM), or erlotinib (10 μM) for 24 hours followed by co-IP of anti-apoptotic members to evaluate for changes in Bim binding. B. U0126 synergizes with ABT-737, supporting ERK as the downstream EGFR effector regulating Mcl-1 dependence in HR NB. C. Noxa expression increases following EGFR inhibition with erlotinib but is unchanged by treatment with the MEK/ERK inhibitor U0126.
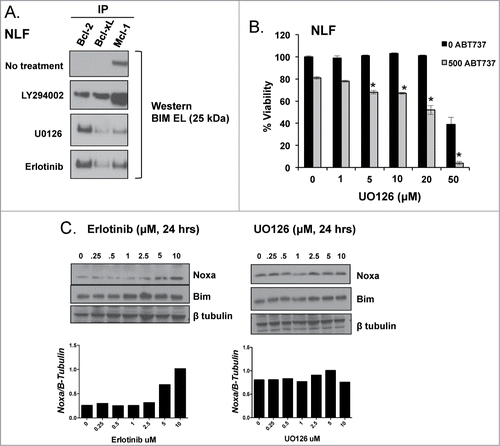
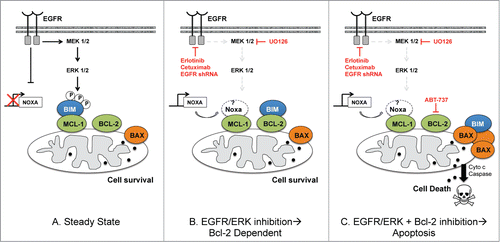
Figure 8. Schema showing EGFR regulation of Bim:Mcl-1 interactions in Mcl-1 dependent HR NB at steady state (left) and how Mcl-1 dependent NB cell survival is affected by molecular/genetic inhibition of EGFR or ERK alone (middle) or in combination with ABT-737 (right). The role of Noxa has not been confirmed by our results and so is left in white.