Figures & data
Figure 1. Identification of ER+ breast cancer specific modules using WGCNA. The clustering dendrogram of gene profilers from the data set GSE6532 with 87 ER+ breast cancer patients. Hierarchical cluster analysis dendrogram was used to detect coexpression clusters. Each short vertical line corresponds to a gene and the branches are expression modules of highly interconnected groups of genes with a color to indicate its module assignment. In total, 9 modules ranging from 37 to 507 genes in size were identified. The gray color suggests the 1546 genes that are outside of all the modules.
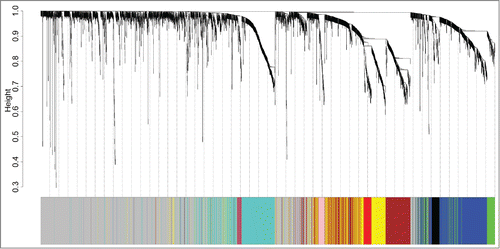
Table 1. Basic characteristics of the datasets
Table 2. Association of expression modules with tumor grade, tumor size, and survival in discovery set
Figure 2. Elevated expression of the turquoise ME, a group of coexpressed genes indicates poor outcome in ER+ breast cancer patients treated with tamoxifen. Kaplan–Meier survival plots for RFS in the training (A) and validating (B) datasets. Increased expression (red) of this coexpressed group is associated with poor RFS. (C) GO enrichment analysis for the 507 genes comprising the turquoise module identifies multiple processes related to cell proliferation. The original significance outputted from DAVID for GO biological processes were transformed in to “–log (P-value)” for plotting.
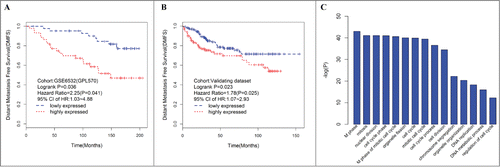
Table 3. Association relationship between hub genes with survival
Table 4. Association relationship between hub genes with survival within breast cancer molecular subtypes
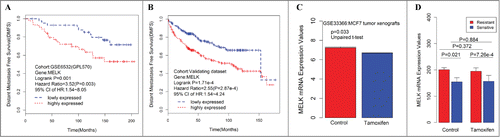
Figure 3. Consistent associations between RFS and the MELK genes are observed across the training data set and validating data set. Kaplan–Meier survival plot for RFS for MELK indicates increased expression (red) of this gene indicates poor prognosis in the training dataset (A) and validating data set (B). Breast cancer patients grouped by the median of gene expression level, significances were assessed by logrank test. (D) Log2 transformed mRNA expression values of MELK in MCF-7 tumor xenografts treated with either tamoxifen or control. P values were calculated by independent 2-tailed t test. Error bars represent mean ± SD. (E) mRNA expression values of MELK in tamoxifen resistant/sensitive MCF-7 subclones treated with tamoxifen or control. P values were calculated by unpaired 2-tailed t test. Error bars represent mean ± SD.