Figures & data
Figure 1. MiR-15a and miR-16 increase autophagic activity. (A, B) MiR-15a and miR-16 promote GFP-LC3 puncta formation. HeLa cells stably expression GFP-LC3 were transfected with miR-15a, miR-16 or negative control (NC). Cells were fixed at 48 h post transfection. (A) Representative images were captured by confocal microscope. Scale bar, 10 μm. (B) Quantitative analysis of GFP-LC3 puncta in (A). Data shown are means ± SD of three independent experiments. **P < 0.01. (C) Over-expression of miR-15a or miR-16 induces LC3-II conversion and p62 degradation. Densitometric analysis were calculated using Image J software. (D) Overexpression of miR-15a and miR-16 increase autophagic flux. HeLa cells transfected with NC, miR-15a or miR-16 were treated with Baf A1 (10 nM) for 2 h. Western blotting was performed to analyze the status of LC3, p62 and GAPDH. (E) Densitometric analysis of LC3-II/GAPDH or (F) p62/GAPDH ratios from immunoblots using Image J. Data shown are means ± SD of three independent experiments. *P < 0.05, **P < 0.01, student 2-tailed t test.
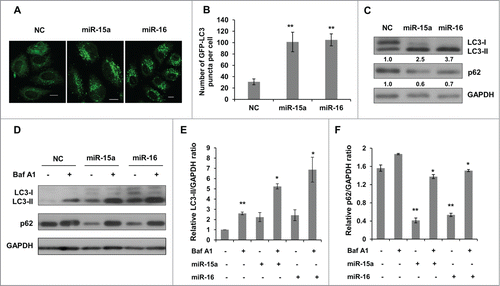
Figure 2. Inhibition of endogenous miR-15a and miR-16 repress autophagic activity. (A) Blockage of endogenous miR-15a or miR-16 expression by transfection of single strand ant-miRNA. (B) Inhibition of endogenous miR-15a or miR-16 inhibits GFP-LC3 puncta formation. HeLa cells transfected with ant-miR-15a, ant-miR-16 or control (ant-NC) were fixed at 48 h after transfection. Representative images were captured by confocal microscope. Scale bar, 10 μm. (C) Quantitative analysis of GFP-LC3 puncta per cell. At least 200 cells were examined in each experimental group. Data shown are means ± SD of three independent experiments. **P < 0.01. (D) HeLa cells were transfected with ant-miR-15a, ant-miR-16 or ant-NC. Western blotting was performed to analyze the status of LC3, p62, and GAPDH.
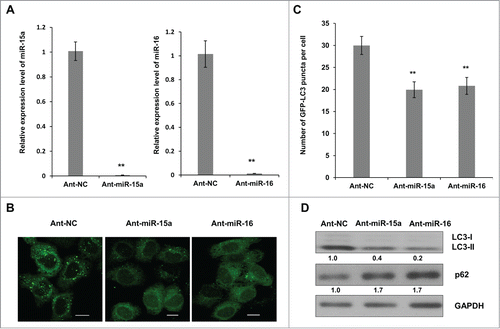
Figure 3. Rictor is a direct target of miR-15a and miR-16. (A) Predicted binding sequence of miR-15a, miR-16 in Rictor 3′ UTR. (B) Western blot analysis of Rictor and GAPDH proteins in HeLa cells transfected with miR-15a, miR-16, NC, ant-miR-15a, ant-miR-16 or anti-NC. Protein ratios were determined by ImageJ densitometric analysis. (C) Luciferase reporter assay of wild type (WT) or mutated (MUT) Rictor 3′UTR vector co-transfected with NC, miR-15a or miR-16 respectively. Data shown are means ± SD of three independent experiments, **P < 0.01, student 2-tailed t test. (D) HeLa cells were transfected with Rictor siRNAs, miR-15a, miR-16 or NC. Western blotting was performed to analyze the status of LC3, p62, Rictor and GAPDH.
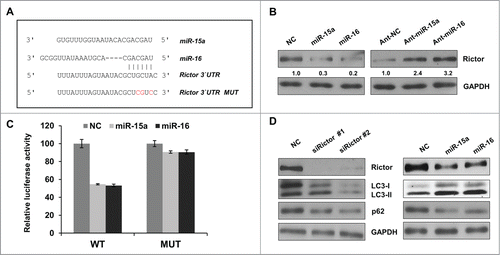
Figure 4. MiR-15a and miR-16 inhibit cell proliferation partly through autophagy. (A) HeLa cells were transfected with Rictor siRNAs, miR-15a, miR-16 or NC. Western blotting were performed to analyze the status of total and phosphorylated mTOR and p70S6K. (B) MTT assay of HeLa cells transfected with miR-15a, miR-16, Rictor siRNA or NC, in the presence or absence of ATG7 siRNA. Cell viability (OD 490 nm absorbance) was examined at 48 h after transfection. Data shown are means ± SD of three independent experiments, **P < 0.01, student 2-tailed t test. (C) miR-15a and miR-16 induces G1/S cell cycle arrest. HeLa cells transfected with miR-15a, miR-16, Rictor siRNA or NC were synchronized at G1/S boundary by treatment with hydroxyurea (HU). Cells were released from HU block for 4 h, fixed and stained with with PI for Flow cytometry analysis. (D) Inhibition of autophagy partially rescues miR-15a and miR-16 induced G1/S cell cycle arrest. HeLa cells co-transfected with ATG7 siRNA and miR-15a, miR-16, Rictor siRNA or NC were synchronized at G1/S and released for 4 h. Percentage of G1, S and G2/M cells was quantified from flow cytometry analysis.
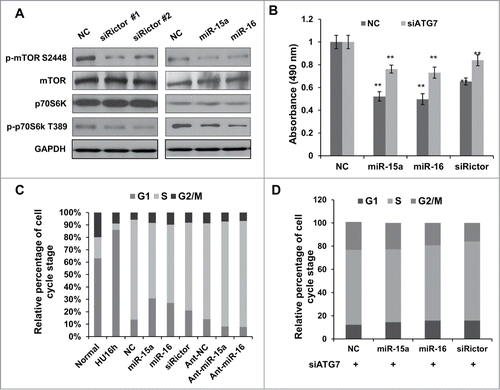
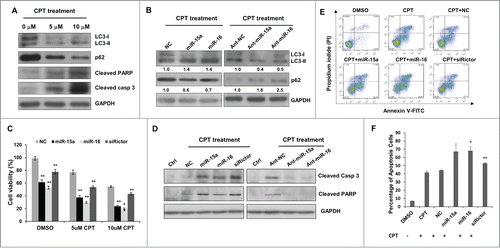
Figure 5. MiR-15a and miR-16 enhance chemotherapeutic efficacy of Camptothecin. (A) Camptothecin (CPT) induced LC3 conversion, p62 degradation, and cleavage of caspase 3 and PARP. (B) Transfection of miR-15a or miR-16 increased CPT-induced autophagy and inhibition of endogenous miR-15a or miR-16 had the opposite effects on CPT-induced autophagy. Image J densitometric analysis of LC3-II/GAPDH and p62/GAPDH was shown. (C) Cell viability assay of HeLa cells transfected with miR-15a, miR-16 or Rictor siRNA in DMSO or CPT treated cells. Data shown are means ± SD of three independent experiments, **P < 0.01, student 2-tailed t test. (D) Western blot analysis of cleaved PARP1 and caspase 3 in HeLa cells transfected with miR-15a, miR-16, Rictor siRNA, NC or ant-miR-15a, ant-miR-16, ant-NC and treated with CPT for 24 h. (E) HeLa cells transfected with miR-15a, miR-16, Rictor siRNA, or NC were incubated with 10 uM CPT for 36 h. Cells were harvested and stained with PtdIns and annexin V-FITC for apoptotic analysis. (F) The percentage of apoptotic cells were the sum of the early apoptotic cells located in the lower right quadrant (annexin V–FITC +/PI - cells), as well as late apoptotic cells located in the upper right quadrant (annexin V–FITC +/PtdIns - cells). Data shown are means ± SD of three independent experiments, *P < 0.05, **P < 0.01, student 2-tailed t test.