Figures & data
Figure 2. Interactions of Zelpolib at the active site of Pol δ. A, Zelpolib forms two hydrogen bonds with dG on the template strand. The catechol moiety stacks against dOC (dideoxycytosine) on the primer. B, comparative position of dCTP at the active site of Pol δ. C, chemical structure of Zelpolib. D, 5 potential hydrogen-bonds can be formed between Zelpolib and Pol δ.
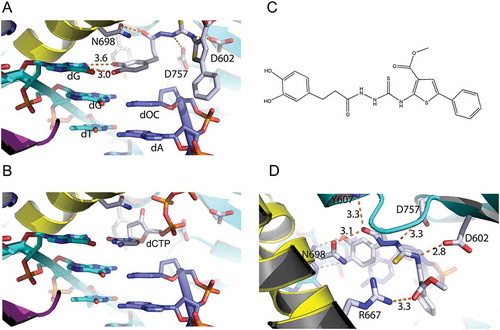
Figure 3. Comparison of Zelpolib with dCTP in binding to the active site of Pol δ. Zelpolib in stick (A) and ball (B) representation with dCTP (C and D) in comparison.
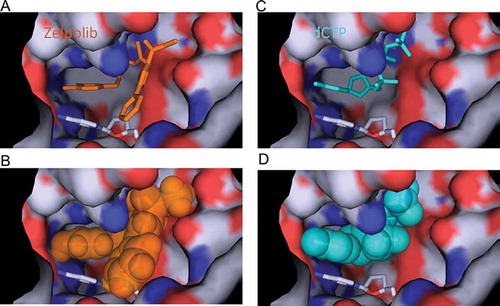
Figure 4. Zelpolb inhibits Pol δ activity. A, enzymatic assay by poly(dA)/oligo(dT) method demonstrates noncompetitive inhibition with Ki of 4.3 μM. B, Zelpolib is likely unique in inhibiting Pol δ activity in comparison to current FDA approved small molecule oncology drugs. C, quantification of the full-length products (40mer, integration by ImageJ). Lower peak height indicates less product. D, quantification of unextended primer. Higher peaks correspond to higher amounts of primer left.
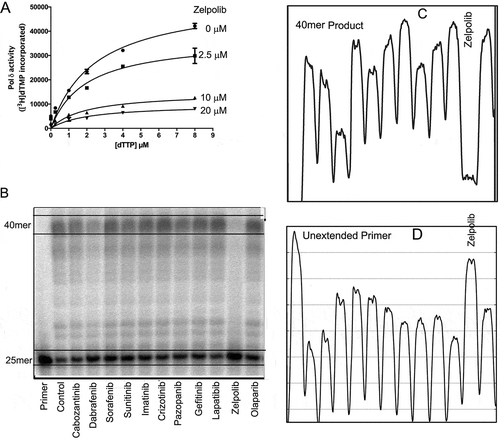
Figure 5. Zelpolib inhibits cellular DNA replication. A, EdU incorporation by whole cell population is inhibited by Zelpolib. Exponentially growing HCC1395 (TNBC) cells were treated with Zelpolib for 2 hours prior to pulse labeling with EdU for 30 minutes. Amount of EdU quantified with “click” chemistry and measured by laser scanning cytometry (LSC). Error bar shows mean value with SEM (triplicates) and P values were calculated using unpaired Student’s t-test. B, treatment scheme of DNA fiber fluorography assay. C, DNA fiber-length comparison between untreated and treated with 50 μM Zelpolib (HCC1395 cells, see Figure S4 for original images). D, quantification of DNA fiber length for BxPC-3 cells. E, quantification of DNA fiber length for HCC1395 cells. 75 fibers were analyzed per sample. Scattered dot plot shows ratio of green/red fiber lengths (ratios) with SEM. P values were calculated using unpaired T test. *** indicates p < 0.0001.
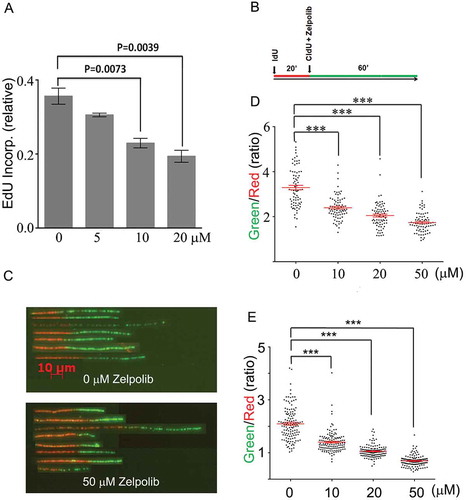
Figure 6. Antiproliferative activities of Zelpolib. Concentration dependent inhibition of cell proliferation by Zelpolib on three different cell lines and comparison with methotrexate, 5-FU, and cisplatin by MTT assays. All samples were in triplicates and presented as averages with standard deviation.
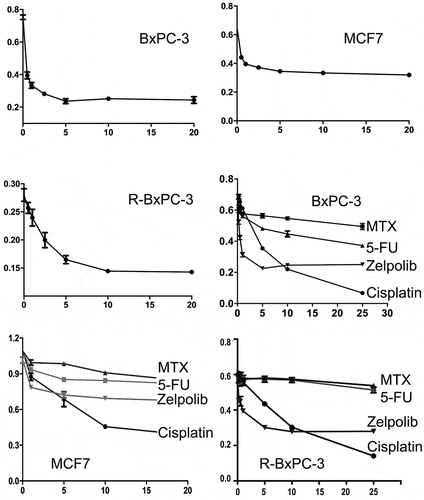
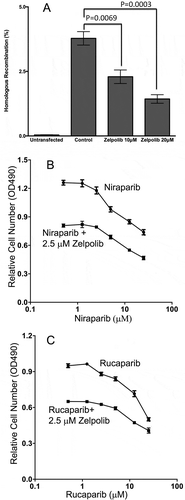
Figure 7. Zelpolib inhibits homologous recombination in cell-based assay. a, Dual plasmids (DR-GFP and I-Scel) reporter assay was used to measure the effect of Zelpolib on DSB repair in 293T cells as reportedCitation34. The error bar represents mean values of four repeats with SEM. P values were calculated using unpaired T test. b, Zelpolib enhances the sensitivity of triple negative breast cancer cells (HCC1395) to niraparib by MTT assay. c, Zelpolib enhanced the sensitivity of TNB cells (HCC1395) towards Rucaparib.