Figures & data
Figure 1. ADCC resistance in multiple cell line models is characterized by loss of NK cell conjugation (a-c), percent cytotoxicity of ADCC-sensitive (ADCCS, blue) compared to ADCC-resistant (ADCCR, red) A431 (a), SKOV3 (bB), and FaDu (c) cells as measured by ADCC assay (n = 3). Unpaired two-tailed t-test, *, P<.05, **, P<.01. Error bars, SEM. D-F, percent of NK92-CD16V cell-target cell conjugates in ADCC-sensitive (ADCCS, blue) compared to ADCC-resistant (ADCCR, red) A431 (d), SKOV3 (e), and FaDu (f) cells when treated with NK92-CD16V cells alone (open bar) or ADCC conditions (full bar) after 2hrs (n = 5 for A431, n = 4 for SKOV3 and FaDu). Unpaired two-tailed t-test, *, P<.05, ns, not significant. Error bars, SEM.
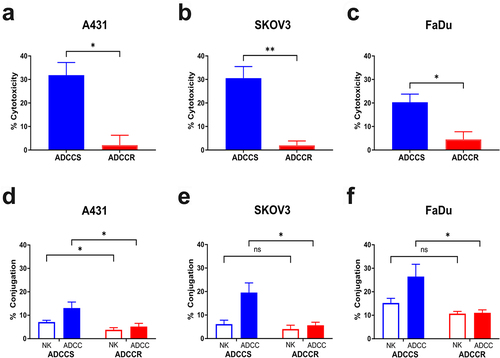
Figure 2. Variable loss of antibody target in ADCC resistance (a), surface EGFR expression measured by flow cytometry in ADCC-sensitive (blue) and ADCC-resistant (red) A431 cells with isotype control staining in ADCC-sensitive (light blue) and ADCC-resistant (light red) cells. Representative histogram from n = 3 experiments. (b), Representative immunofluorescence images of ADCC-sensitive (ADCCS) compared to ADCC-resistant (ADCCR) A431 cells incubated with Dylight550-conjugated cetuximab for 4hrs; red: cetuximab and blue: DAPI. Representative of n = 2, magnification 40×, scale bar = 25 μm. (c), surface HER2 expression measured by flow cytometry in ADCC-sensitive (blue) and ADCC-resistant (red) SKOV3 cells with isotype control staining in ADCC-sensitive (light blue) and ADCC-resistant (light red) cells. Representative histogram from n = 3 experiments. (d), Representative immunofluorescence images of ADCC-sensitive (ADCCS) compared to ADCC-resistant (ADCCR) SKOV3 cells incubated with Dylight550-conjugated trastuzumab for 3hrs; red: trastuzumab and blue: DAPI. Representative of n = 2, magnification 40×, scale bar = 25 μm. (e), surface EGFR expression measured by flow cytometry in ADCC-sensitive (blue) and ADCC-resistant (red) FaDu cells with isotype control staining in ADCC-sensitive (light blue) and ADCC-resistant (light red) cells. Representative histogram from n = 3 experiments. (f), Representative immunofluorescence images of ADCC-sensitive (ADCCS) compared to ADCC-resistant (ADCCR) FaDu cells incubated with Dylight550-conjugated cetuximab for 3hrs; red: cetuximab and blue: DAPI. Representative of n = 2, magnification 40×, scale bar = 25 μm.
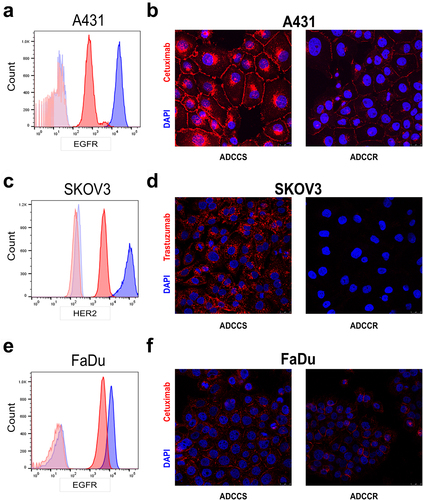
Figure 3. ADCC resistance is associated with loss of cell surface proteins involved in immune synapse formation (a-d), geometric mean of CD95 (FasR) (a), CD262 (TRAIL-R2/DR5) (b), CD104 (Integrin beta 4) (c), and CD49f (Integrin Alpha 6) (d) expression as measured by BioLegend LEGENDScreen™ in ADCC-sensitive (blue) compared to ADCC-resistant (red) cells. (e), percent cytotoxicity of each ADCC-sensitive and ADCC-resistant cell line as measured by ADCC assay when untreated (white bar), pretreated with a combination of 10 µg/ml of each isotype control antibody (gray bar), or pretreated with a combination of 10 µg/ml of each blocking antibody for CD95, CD262, CD104, and CD49f (black bar) for 1 hr (n = 4). Unpaired two-tailed t-test, ns, not significant. Error bars, SEM. F, percent cytotoxicity of each ADCC-sensitive and ADCC-resistant cell line as measured by ADCC assay when untreated (white bar), pretreated with 10 µg/ml of isotype control antibody (gray bar), or pretreated with 10 µg/ml of blocking antibody for CD54 (black bar) for 1 hr (n = 3). Unpaired two-tailed t-test, *, P<.05, ns, not significant. Error bars, SEM. G, Representative flow cytometry analysis of CD54 (ICAM1) in ADCC-sensitive (blue) and ADCC-resistant (red) A431 cells with isotype control staining in ADCC-sensitive (light blue) and ADCC-resistant (light red) cells.
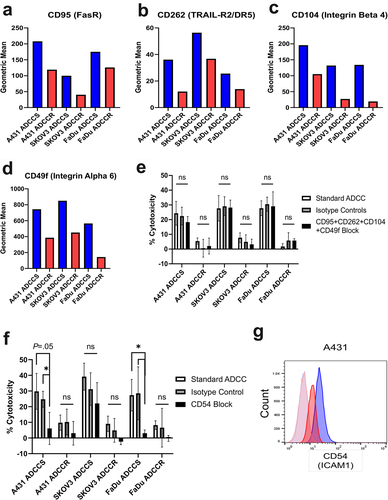
Figure 4. Single-cell CITE-seq identifies ADCC resistance associated genes A-C, volcano plots of genes differentially expressed between A431 (a), SKOV3 (b), and FaDu (c) ADCC-sensitive and ADCC-resistant cells. P-value was calculated by Wilcoxon rank-sum test and then adjusted by Bonferroni correction. Genes with – Log10(p-value)>1.3 are marked in blue and genes with both – Log10(p-value)>1.3 and log2 fold change (FC) ±0.5 were considered significant and are marked in red. NS, not significant. D, violin plots of the gene expression level in each cell line for the six genes with shared statistically significant upregulation in all ADCC-resistant cell lines. E and F, violin plot of EGFR (E) and ERBB2 (F) gene expression level in each ADCC-sensitive and -resistant cell line.
Figure 5. STAT1 signaling is associated with but does not drive acquisition of ADCC resistance (a), western blot analysis of STAT1 pathway proteins in ADCC-sensitive (ADCCS) and ADCC-resistant (ADCCR) A431, SKOV3, and FaDu cells. Densitometry values for expression normalized to GAPDH in ADCC-resistant relative to ADCC- sensitive cells are indicated. (b), percent cytotoxicity of ADCC-sensitive (blue) and ADCC-resistant (red) A431 cells as measured by ADCC assay when cells are incubated in either medium alone (solid bars) or medium supplemented with DMSO (horizontal hash bars) or 10 nM ruxolitinib (diagonal hash bars) for 72hrs prior to ADCC assay (n = 3). Unpaired two-tailed t-test, ns, not significant. Error bars, SEM. (c), Representative western blot analysis of phospho-STAT1 protein expression in ADCC-sensitive and ADCC-resistant A431 cells when cells are incubated in either medium alone (untreated) or medium supplemented with DMSO or 10 nM ruxolitinib for 72hrs. Densitometry values for expression normalized to GAPDH in cells treated with DMSO or ruxolitinib compared to untreated cells are indicated. (d), percent cytotoxicity of ADCC-sensitive (blue) and ADCC-resistant (red) A431 cells as measured by ADCC assay when cells are incubated in either medium alone (solid bars) or transfected with control scramble siRNA (siNEG, horizontal hash bars) or siRNA specific for STAT1 (diagonal hash bars) for 48 h prior to ADCC assay (n = 3). Unpaired two-tailed t-test, **, P<.01. Error bars, SEM. (e), Representative western blot analysis of STAT1 protein expression in ADCC-sensitive and ADCC-resistant A431 cells when cells are incubated in either medium alone (untreated) or transfected with control scramble siRNA (siNEG) or siRNA specific for STAT1 for 48hrs. Densitometry values for expression normalized to GAPDH in cells transfected with control scramble (siNEG) or siRNA specific for STAT1 compared to untreated cells are indicated. (f), western blot analysis of STAT1 protein expression in untreated and ADCC condition treated parental A431 cells, A431 cells transduced with a non-targeting control CRISPR-cas9 (NTC), and A431 cells transduced with CRISPR-cas9 specific for STAT1 (STAT1-KO) at the indicated challenges. (g), percent cytotoxicity of untreated and ADCC condition treated parental A431 cells, A431 cells transduced with a non-targeting control CRISPR-cas9 (NTC), and A431 cells transduced with CRISPR-cas9 specific for STAT1 (STAT1-KO) at every 5 challenges during derivation of resistance as measured by ADCC assay (n = 2). Unpaired two-tailed t-test, *, P<.05. Error bars, SEM.
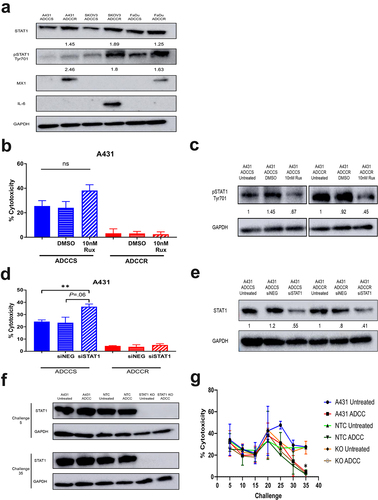
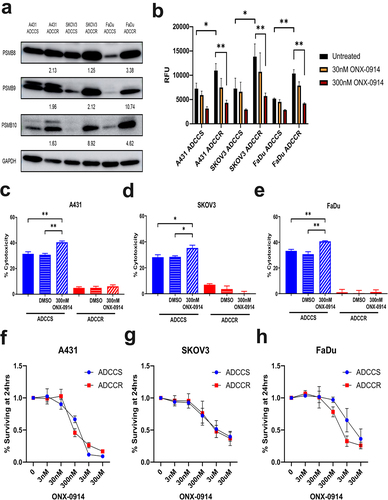
Figure 6. Immunoproteasome activation is associated with ADCC resistance (a), western blot analysis of immunoproteasome subunit expression in ADCC-sensitive (ADCCS) and ADCC-resistant (ADCCR) A431, SKOV3, and FaDu cells. Densitometry values for expression normalized to GAPDH in ADCC resistant relative to ADCC sensitive cells are indicated. (b), immunoproteasome activity in ADCC-sensitive (ADCCS) and ADCC-resistant (ADCCR) A431, SKOV3, and FaDu cells as measured by fluorometric plate based assay. Relative fluorescent units (RFU) were measured for cleavage of immunoproteasome specific substrate to 7-amino-4-methylcoumarin after 1 hr in the presence of absence of the immunoproteasome inhibitor ONX-0914. C-E, percent cytotoxicity of ADCC-sensitive (blue) and ADCC-resistant (red) A431 (c), SKOV3 (d), and FaDu (e) cells as measured by ADCC assay when cells are incubated in either medium alone (solid bars) or medium supplemented with DMSO (horizontal hash bars) or 300 nM of the immunoproteasome inhibitor ONX-0914 (diagonal hash bars) for 1 hr prior to ADCC assay (n = 3 for A431 and FaDu, n = 4 for SKOV3). Unpaired two-tailed t-test, *, P<.05, **, P<.01. Error bars, SEM. F-H, cell viability represented as the percent of cells remaining relative to the untreated ADCC-sensitive (blue) and ADCC-resistant (red) A431 (f), SKOV3 (g), and FaDu (h) cells as measured by crystal violet staining of cells after 24hrs of incubation in either medium alone (untreated) or medium supplemented with various concentrations of ONX-0914.
Supplemental Material
Download MS Word (57.7 KB)Supplemental Material
Download MS Word (22.3 KB)Supplemental Material
Download MS Word (11 MB)Data availability statement
The data that support the findings of this study are in GEO, accession number GSE206955 and are available upon request from the corresponding author, LMW.
