Figures & data
Figure 1. LINC00839 is deregulated in the bladder cancer.
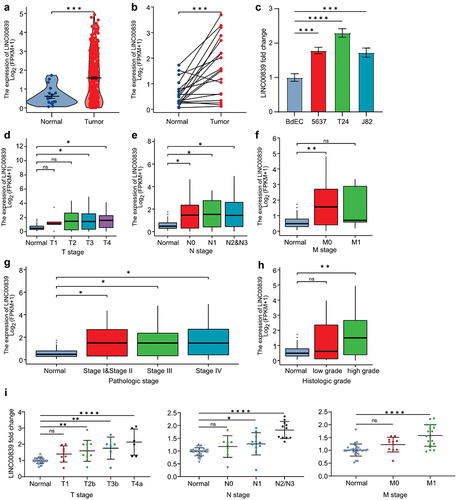
Figure 2. LINC00839 promotes the migration, invasion and EMT of bladder cancer cells.
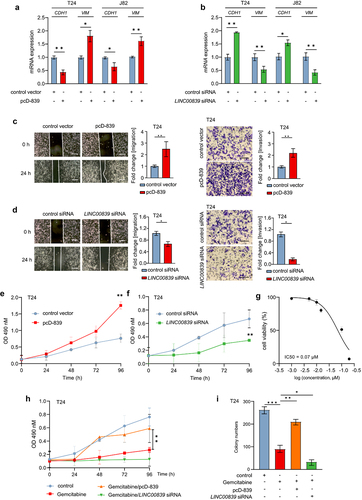
Figure 3. LINC00839 is a direct target of EGR1.
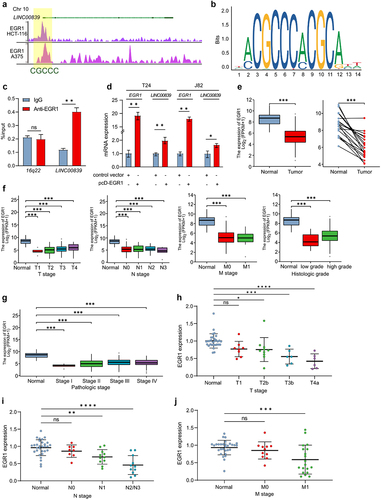
Figure 4. EGR1 represses migration and invasion of bladder cancer cells.
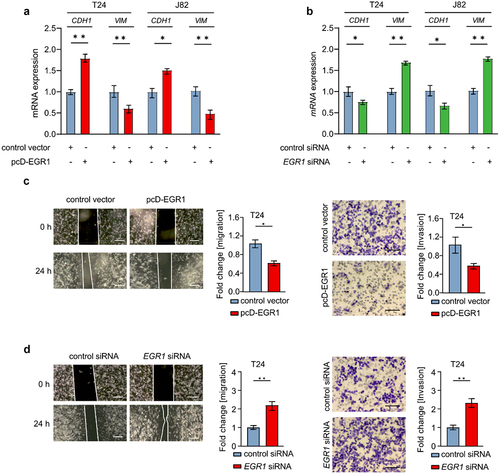
Figure 5. LINC00839 interacts with miR-142.
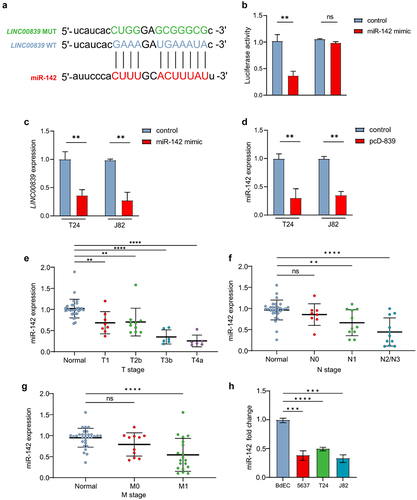
Figure 6. LINC00839/miR-142 axis modulates the expression of SOX5.
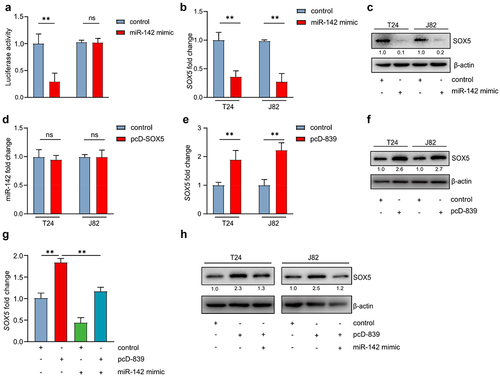
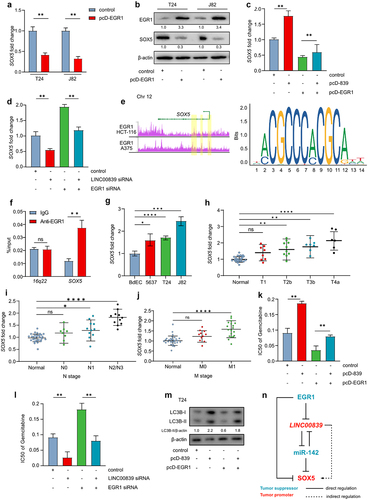
Figure 7. EGR1 suppresses SOX5 via a coherent feed-forward loop.