Figures & data
Figure 1. P4HA2 expression is upregulated in CRC tumor tissues and related bad clinical outcomes.
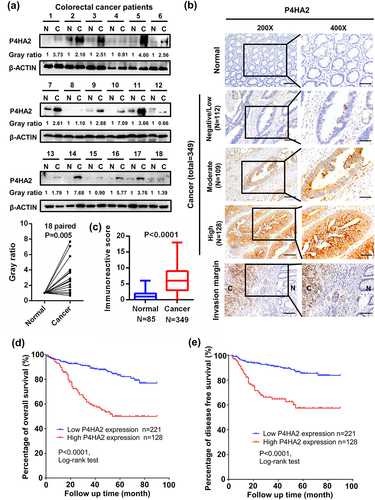
Table 1. Baseline characteristics of patients.
Table 2. Comparison between P4HA2 high and low cases.
Table 3. Relationship between clinicopathological characteristics and OS/DFS.
Figure 2. P4HA2 promotes CRC cell proliferation by regulating cell cycle in vivo and in vitro.
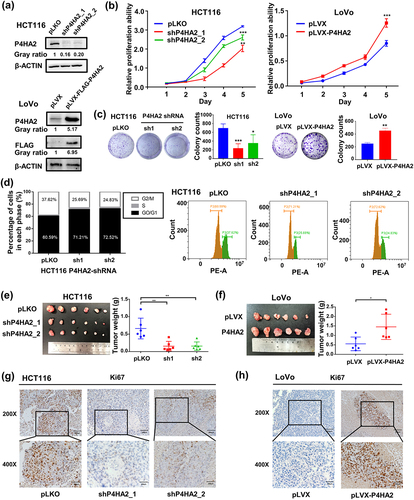
Figure 3. P4HA2 facilitates tumor migration by inducing EMT.
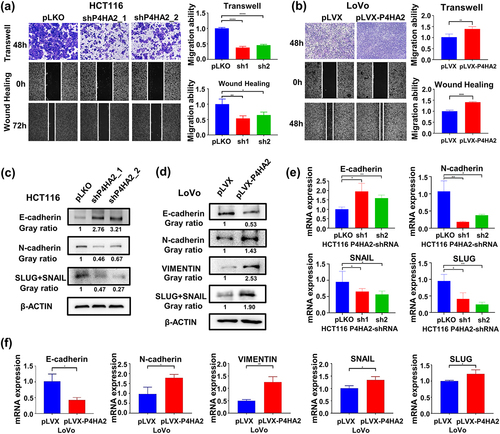
Figure 4. AGO1 mediates the effects of P4HA2 on promoting colorectal cancer progression.
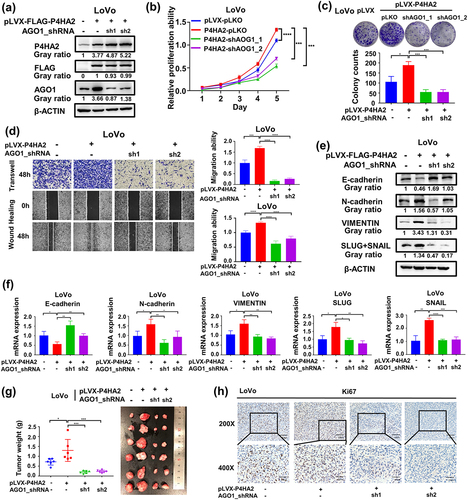
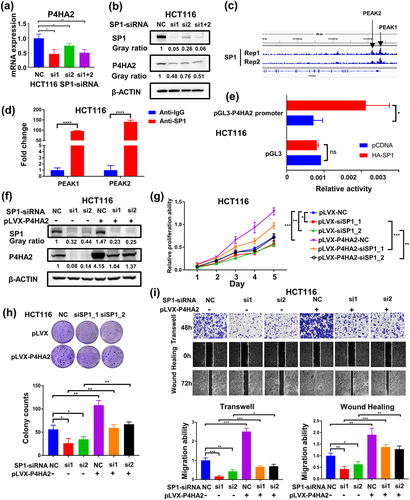
Figure 5. SP1 transcriptionally activates P4HA2 gene expression in colorectal cancer.
Supplementary Table S2 .docx
Download MS Word (17.8 KB)Supplementary Table S1 .docx
Download MS Word (14.8 KB)Data availability statement
The datasets generated during and analyzed during the current study are available from the corresponding author on reasonable request
