Figures & data
Figure 1. MMSET is regulated in a cell-cycle-dependent manner and degraded by the ubiquitin-proteasome pathway during S phase. (A) Asynchronous U2OS cells or cells synchronized at the G1-S border were released into S phase for the indicated times. Lysates were collected for each time point and blotted for the indicated proteins. (B) Asynchronous U2OS cells were plated on coverslips, fixed, and stained for either Cyclin E1 (red) or MMSET (green). Nuclei were visualized with 4',6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI, blue). Scale bar=10 μm. (C) U2OS cells were synchronized at the G1-S border by double-thymidine block and released into S phase for the indicated times either in the absence (top) or presence (bottom) of MG132. Lysates were collected at each time point and blotted for the indicated proteins. (D) His-pulldowns were performed under denaturing conditions using lysates from HEK293 cells transfected with the indicated plasmids and synchronized in S phase using the serum starvation and release method. Purified species were subjected to SDS-PAGE and immunoblotted with the indicated antibodies. Arrowhead and a bar indicate monoubiquitinated and polyubiquitinated MMSET, respectively. Data shown are representative of 3 independent experiments.
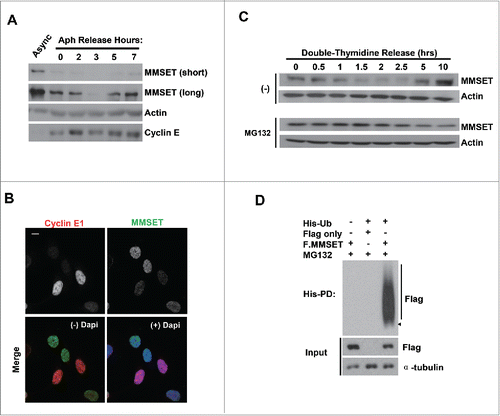
Figure 2. MMSET is degraded by CRL4Cdt2. (A) HEK293T cells were transfected with the plasmids denoted above each lane. Lysates were prepared 48 hours post transfection, immunoprecipitated with anti-Flag, and examined for MMSET. Arrowheads denote Flag-YFP (lower) and Flag-Cdt2 (upper). (B) HEK293T cells were transfected with the plasmids denoted above each lane and synchronized in S phase by the serum starvation and release method. Lysates were prepared and examined for the indicated proteins. The numbers beneath the blots provide the densitometric ratio of MMSET signal to Actin, setting the value for the control group as 1. (C) HEK293T cells were transfected as in B, fixed, and stained for either Cdt2 (Flag, red) or MMSET (green). Nuclei were visualized with 4',6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI, blue). Scale bar= 10 μm. (D) HEK293T cells were treated with the indicated siRNAs for 24 hours, followed by an aphidicolin block to synchronize cells into S phase. Cells were released into S phase for 4 hours in the presence of cycloheximide and lysates were prepared and examined for the indicated proteins. The numbers beneath the blots provide densitometric ratio of MMSET or Cdt2 signal to Tubulin, setting the value for the control group as 1. *Denotes a non-specific band detected by the Cdt2 antibody. (E) His-pull-downs were performed under denaturing conditions using lysates from HEK293 cells transfected with the indicated plasmids and synchronized in S phase using the serum starvation and release method. Purified species were subjected to SDS-PAGE and immunoblotted with the indicated antibodies. Arrowhead and a bar indicate monoubiquitinated and polyubiquitinated MMSET, respectively. (F) His-pull-downs were performed under denaturing conditions using lysates from HEK293 cells transfected with the indicated plasmids and synchronized in S phase using the serum starvation and release method. Purified species were subjected to SDS-PAGE and immunoblotted with the indicated antibodies. Arrowhead and a bar indicate monoubiquitinated and polyubiquitinated MMSET, respectively. Data shown are representative of 3 independent experiments.
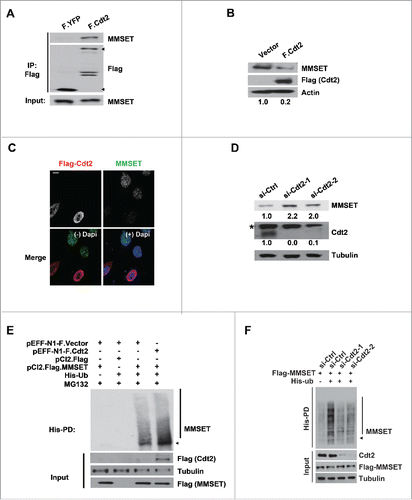
Figure 3. MMSET interacts with PCNA via its N-terminus and PCNA depletion promotes MMSET stability. (A) U2OS cells were immunoprecipitated with PCNA or mouse IgG and the resulting samples were examined for MMSET or PCNA by western blot. (B) Left: T98G cells were transfected with the indicated siRNAs and synchronized in S phase by the serum starvation and release method. Lysates were prepared 48 hours later and examined for the indicated proteins. Right: cell-cycle profiles of the samples. (C) Schematic representation of MMSET fragments used for the GST pull-down assay. (D) A GST-pull down assay was carried out between purified PCNA and GST-MMSET fragments. Coomassie blue staining shows GST fusion proteins used in the pull-down assay (middle). Data shown are representative of 3 independent experiments.
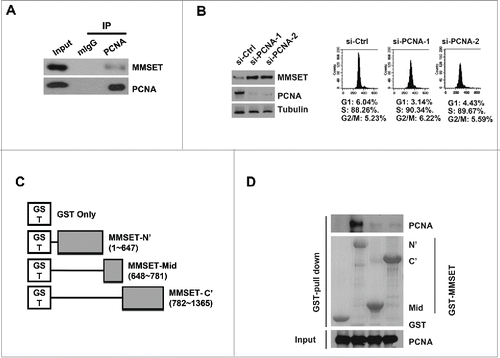
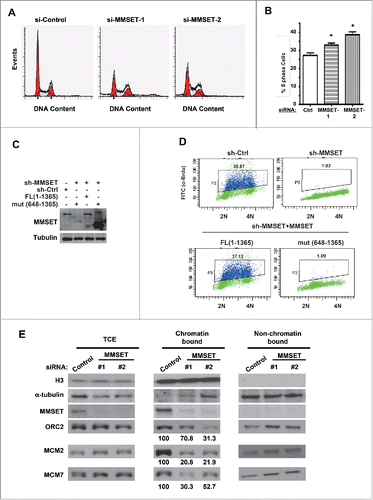
Figure 4. MMSET is required for normal cell-cycle progression. (A) HeLa cells were treated with the indicated siRNAs and, 48 hours later, cells were fixed and examined for DNA content by flow cytometry. (B) Cell-cycle profiles from A were analyzed and S phase was plotted for each condition. Error bars indicate SEM. *P < 0.05. (C) HCT116 cells stably expressing MMSET shRNA were reconstituted with WT MMSET or the MMSET deletion mutant (648-1356) and the indicated proteins were examined by western blot. (D) A subset of cells from C was examined for DNA content and BrdU incorporation using flow cytometry. (E) HeLa cells were treated with the indicated siRNAs, synchronized at the G1-S border using aphidicolin, and total cell extracts (TCE) or chromatin and non-chromatin enriched fractionations were prepared 48 hours post-transfection. The indicated proteins were examined by western blot. The numbers beneath the blots provide the densitometric ratio of ORC2, MCM2, or MCM7 signal to H3, setting the value for the control group as 100. Data shown are representative (A, C, D, and E) or an average of 3 independent experiments (B).