Figures & data
Figure 1. PrimPol-deficient cells are sensitive to a wide range of DNA damaging agents. PrimPol −/− cells exhibit hypersensitivity to various types of DNA damage. Cells with the indicated genotype were exposed to the indicated genotoxic agents. The dose of the genotoxic agent is displayed on the x-axis on a linear scale, while the percent fraction of surviving cells is displayed on the y-axis on a logarithmic scale. Error bars show the SD of the mean for 3 independent assays.
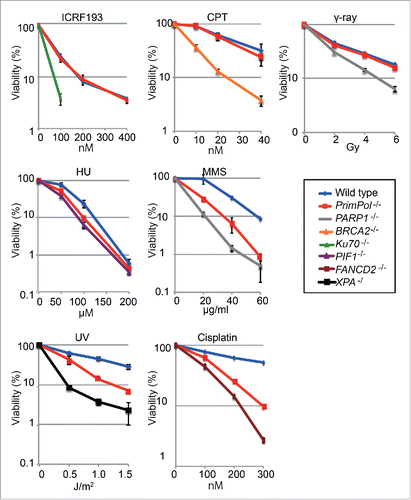
Figure 2. PrimPol plays roles in damage tolerance independently of Polη and Polζ. (A) Relative growth rate of cells plotted with indicated genotypes. Doubling time for the indicated cells was calculated. Error bars represent standard deviation from independent experiments (n = 3). (B) Indicated cells were treated with 0 or 100 nM of cisplatin for 16 hr. Representative cell-cycle distribution for the indicated genotypes. The top of the box, and the lower left, lower right, and left-most gates correspond to cells in the S, G1, and G2/M phases, and the sub-G1 fraction, respectively. The sub-G1 fraction represents dying and dead cells. The percentage of cells in each gate is indicated. (C) Percentage of the indicated cells in sub-G1 fraction and G2 phase fraction was indicated. Error bar represent standard deviation from independent experiments (n = 3). Statistical significance was determined by a Student's t-test and p-value was calculated. (*) p < 0.05 (D) Indicated cells were exposed to UV or cisplatin and sensitivities were indicated as in .
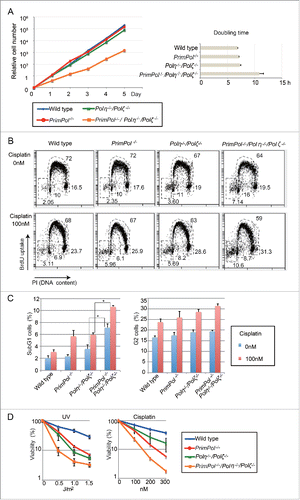
Figure 3. PrimPol is dispensable for TLS past abasic sites during Ig Vλ hypermutation. (A) Ig Vλ segments isolated from indicated cells, clonally expanded for two weeks. Horizontal lines represent the rearranged Ig Vλ (450 bp), with hypermutation (lollipop shapes), gene conversion (horizontal bars), single-nucleotide substitutions that could be the result of hypermutation or gene conversion (vertical bars), and single-base deletion (boxes) determined as described previously.Citation3,5,29,31,32 More than three clones were expanded for two weeks and analyzed for each data set. (B) Rate of gene conversion (GC) and hypermutation (PM) events are indicated with standard error. Statistical significance was determined by a Student's t-test and p-value was calculated. (C) Pattern of point mutation in wild-type, PrimPol−/−, Polη−/−/Polζ−/− and PrimPol−/−/Polη−/−/Polζ−/− cells. Tables show the pattern of mutation in each line.
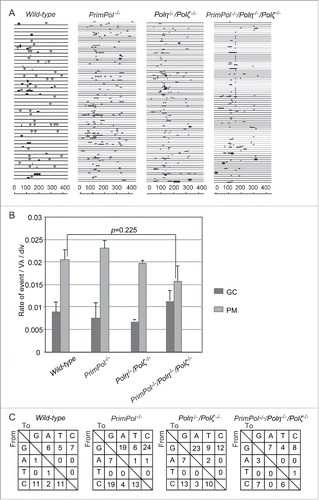
Figure 4. PrimPol's primase activity is required for replication block tolerance. (A) Domain architecture of PrimPol. PrimPol is composed of an AEP polymerase and a Zn2+ finger domain. The PrimPol-Y89D mutation is located in the AEP polymerase. The PrimPol1-354 deletion mutant contains the polymerase domain but lacks the Zn2+ finger domain. A Zn2+ finger domain knockout mutant was generated by mutating the first conserved cysteine and histidine residues that coordinate Zn2+ (C419A and H426A, respectively). (B) PrimPol −/− cells were complemented with variants of PrimPol and their in vivo expression was confirmed by protein gel blot. Asterisks indicate non-specific bands. (C) Cells with the indicated genotype were exposed to the indicated genotoxic agents. The dose of the genotoxic agent is displayed on the x-axis on a linear scale, while the percent fraction of surviving cells is displayed on the y-axis on a logarithmic scale. Error bars show the SD of the mean for three independent assays. (D) Number of the chromosomal aberrations in 100 mitotic cells was presented. DT40 cells were exposed to cisplatin (150 nM) for 14.5 h and colcemid was added 2.5 h before harvest to accumulate a mitotic fraction. Error bars represent SD of the mean for three independent assays. Statistical significance was determined by a Student's t-test and p-value was calculated. (*) p < 0.05 (E) Sensitivity to cisplatin for indicated cells were indicated as in C.
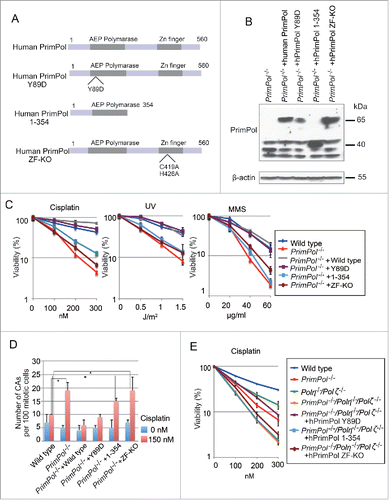
Figure 5. Role of PrimPol's primase activity in cellular tolerance to CTNAs. (A) Cells with the indicated genotype were exposed to the indicated CTNAs. The dose of the genotoxic agent is displayed on the x-axis on a linear scale, while the percent fraction of surviving cells is displayed on the y-axis on a logarithmic scale. Error bars show the SD of the mean for three independent assays. (B) Sensitivity to ACV for indicated cells were indicated as in A.
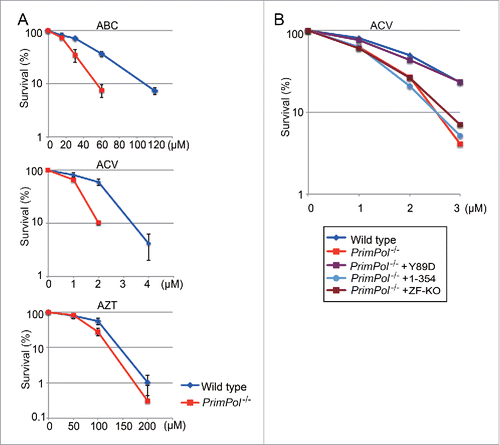
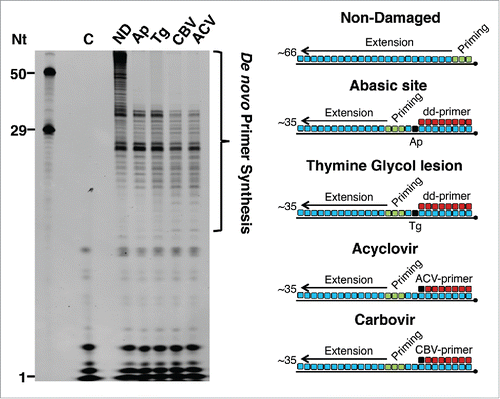
Figure 6. PrimPol catalyzes repriming downstream of 3′ incorporated CTNAs and templating abasic or thymine glycol lesions. PrimPol (1μM) was incubated for 15 min at 37° C with dNTPs (250 μM), FAM-dNTPs (dATP, dCTP, dUTP) (2.5 μM), and mixed sequence primer-templates (1 μM) (as shown in the schematic). Primers containing a 3′ dideoxynucleotide were annealed upstream of the lesion on templates containing a single Ap site (Ap) or thymine glycol (Tg) to allow us to assay for repriming, rather than TLS, activity. In the case of CTNAs, a single CTNA (acyclovir (ACV) or carbovir (CBV)) was located at the 3′ end of the primer in place of the dideoxnucleotide. The length of primase reaction products extended to the end of the template allows analysis of the priming location by PrimPol; the near identical extension products produced in each case show close-coupled repriming by PrimPol downstream of the lesion or CTNA. Oligonucleotide nucleotide (Nt) length markers are shown in the left panel. Priming and extension are represented in the schematic as green and blue, respectively. “C” indicates the no enzyme control. “ND” indicates the non-damaged template without an annealed primer.